

Background: Lupus nephritis (LN) is a severe presentation of systemic lupus erythematosus (SLE). It may affect up to 60% of patients with SLE comprising an important cause of morbidity and mortality [1]. Renal biopsy is the gold standard for diagnosis and severity assessment of renal involvement. However, the invasiveness of kidney biopsy, combined with the fact that early renal involvement can be subclinical, complicates the efforts for early detection and intervention [2]. Non-invasive biomarkers are required for the identification of early-stage LN and form one of the cornerstones for good therapeutic decision-making. Several data are merged in this work, increasing the reliability of the analysis and offering a comprehensive overview of the disease.
Objectives: This study aimed to identify candidate diagnostic tools or targets for LN analyzing transcriptomic datasets.
Methods: We systematically identified and evaluated transcriptomic datasets. GSE11909, GSE50635, GSE110174, GSE154851, GSE32591, and GSE112943 were recuperated from the Gene Expression Omnibus platform, containing transcriptomic profiles of whole blood and renal biopsies from patients with SLE. Differentially Expressed Genes (DEG) were filtered according to the following threshold values: False Discovery Rate (FDR) < 0.05 and log2fc > 2. Metascape was employed in the functional enrichment analysis of these DEGs. Hub genes (HGs) were detected in blood and kidney samples, and the diagnostic potential of the overlapping HGs were screened in dataset GSE99967 drawing receiver operating characteristic (ROC) curves. This dataset contains the transcriptomic profile of SLE patients who had active (ALN) or inactive LN (ANLN) and healthy controls (HC). The proportion of immune cells in whole blood was analyzed by two-way ANOVA with Benjamini-Hochberg correction. Significance was set at p < 0.05.
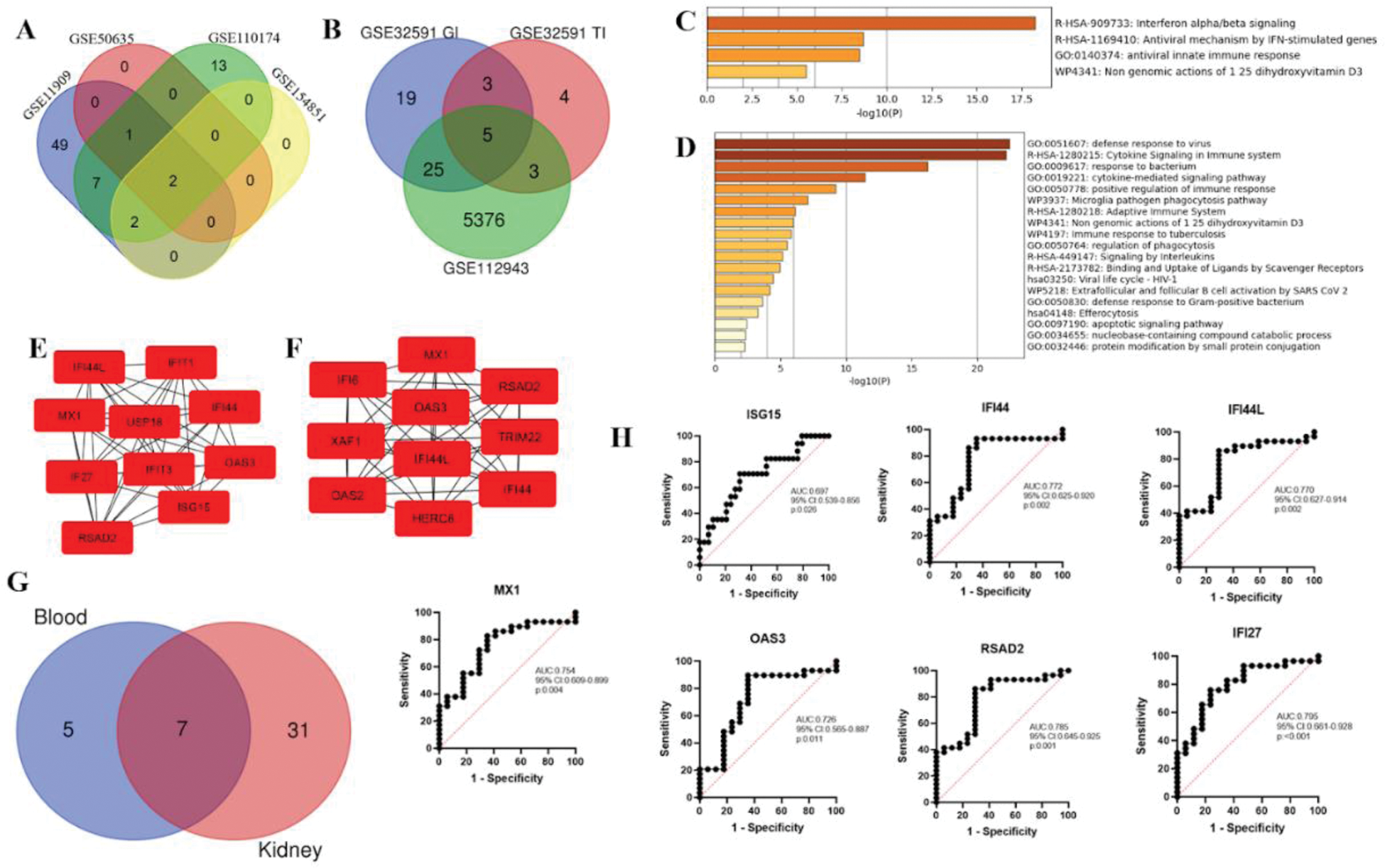
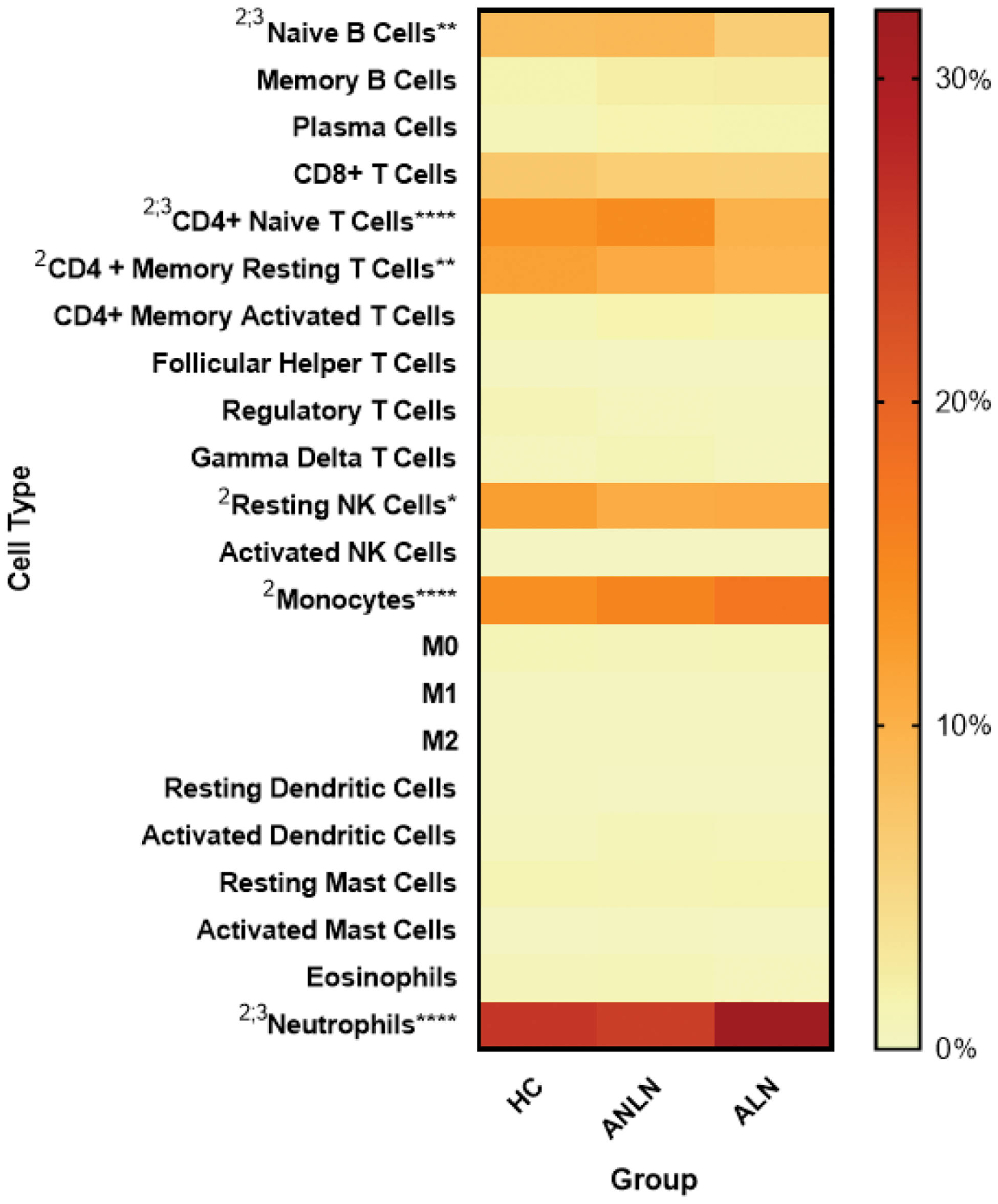
Results: 12 DEGs were identified in blood and 36 in renal tissue. A Venn diagram (Figure 1A, 1B) revealed 7 shared genes between blood and kidney samples. Functional enrichment analysis (Figure 1C, 1D) highlighted key biological processes. HGs such as IFI27 , ISG15 , IFI44 , MX1 , OAS3 , and RSAD2 were identified across blood and kidney samples (Figure 1E–1G), indicating their potential importance in the pathogenesis of LN. ROC curves for these hub genes demonstrated strong diagnostic potential, with area under the curve (AUC) values ranging from 0.72 to 0.79 (Figure 1H). The analysis of immune cell proportions (Figure 2) revealed significant differences in immune cell fractions across the three groups. Notably, increased proportions of neutrophils, CD4+ naïve T cells, and monocytes were observed in the ALN group compared to HC and ANLN. Significant differences were also observed for CD4+ memory resting T cells and resting NK cells.
Differentially expressed genes, hub gene networks, and diagnostic potential in active lupus nephritis. (A) Venn diagram showing the overlap of differentially expressed genes identified in peripheral blood among the datasets. (B) Venn diagram showing overlap of DEGs identified in renal tissue. (C-D) Functional enrichment analysis of the up-regulated DEGs in blood and kidney samples, involved mainly in interferon and cytokine signaling and defense response to virus. Network of the HGs present in blood (E) and kidney (F). (G) Overlapping hub genes between blood and kidney samples presented using a Venn diagram. (H) ROC curves of seven representative HGs, which reflected their diagnostic value for active lupus nephritis. The AUC values ranged from 0.72 to 0.79, reflecting a good predictive power of these biomarkers.
Immune Cell Proportions in Active Lupus Nephritis . Heatmap depicting relative fractions of immune cell types [LM22] across the three groups: healthy control (HC), active non-lupus nephritis (ANLN), and active lupus nephritis (ALN). Significant differences of cell fractions between groups for naïve B cells, CD4+ naïve T cells, CD4+ memory resting T cells, monocytes, resting NK cells, and neutrophils are shown. Comparisons between groups by Two-way ANOVA include HC vs. ANLN (1), HC vs. ALN (2), and ANLN vs. ALN (3). Significance is depicted as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Conclusion: This study identified important genes differentially expressed in peripheral blood and renal tissue of patients with active lupus nephritis, as well as differences in immune cell fractions. These results underline the need for combined transcriptomic and immunological profiling to further elucidate LN pathophysiology, with the goal of developing improved, non-invasive diagnostic modalities.
REFERENCES: [1] Almaani, S., Meara, A., & Rovin, B. H. (2017). Update on lupus nephritis. Clinical Journal of the American Society of Nephrology, 12(5), 825-835. doi:10.2215/CJN.05780616.
[2] Bertsias, G. K., Tektonidou, M., Amoura, Z., Aringer, M., Bajema, I., Berden, J. H., & Boumpas, D. T. (2012). Joint European League Against Rheumatism and European Renal Association–European Dialysis and Transplant Association (EULAR/ERA-EDTA) recommendations for the management of adult and paediatric lupus nephritis. Annals of the Rheumatic Diseases, 71(11), 1771-1782. doi:10.1136/annrheumdis-2012-201940.
Acknowledgements: We’d like to thank the Mexican National Council of Humanities, Science and Technology (CONAHCYT) for providing the academic grant that made this research possible. Grant ID: PCC-2022-320697.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (