

Background: Recent advancements in deep immunophenotyping and serum proteomics have significantly contributed to understand of disease mechanisms. However, studies exploring cross-disease validations across rheumatic and musculoskeletal diseases (RMDs) remain limited, and their potential to uncover disease-specific pathophysiological insights and identify biomarkers that could improve diagnostic precision.
Objectives: This study aimed to investigate pathophysiological variations across RMDs by integrating cellular and protein-level markers and to identify potential biomarkers that may serve as a foundation for future diagnostic strategies.
Methods: Peripheral blood samples were collected from 136 participants, including patients with various RMDs and healthy controls. CyTOF analysis of whole blood, utilizing 37 antibodies, generated 1,272 features, and Olink proteomics quantified 354 serum proteins. Principal component analysis (PCA) was used for unsupervised clustering, and random forest analysis was applied for supervised disease classification. Hyperparameters of the random forest model were optimized using grid research. Data processing and analyses were conducted using Python 3.10.
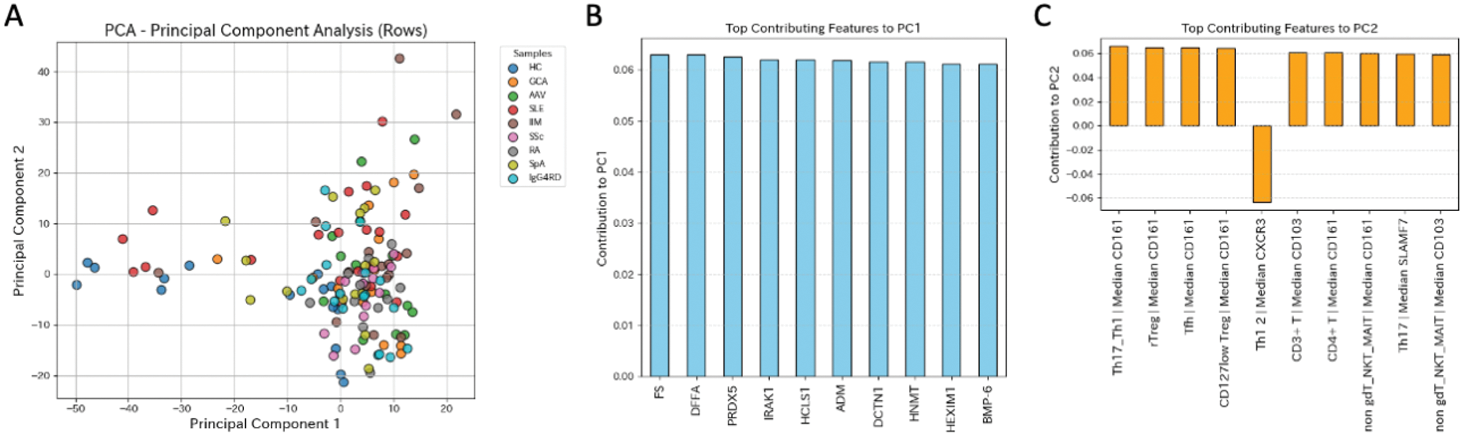
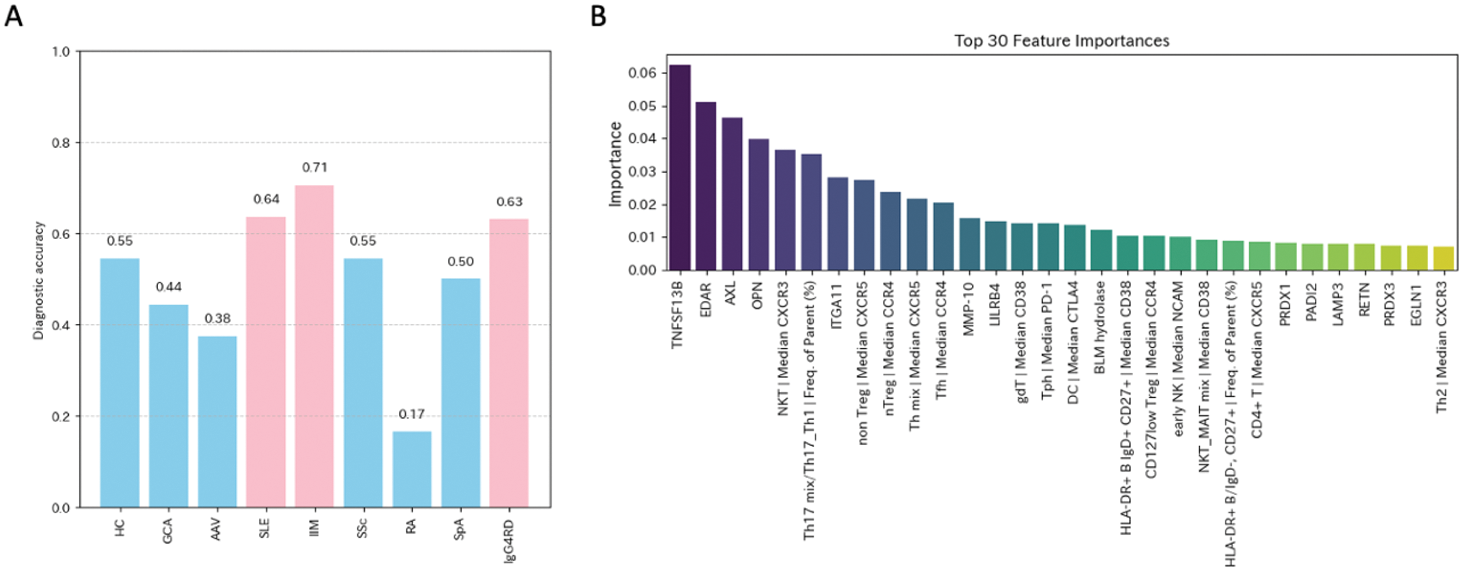
Results: PCA analysis of the combined CyTOF and Olink proteomics datasets revealed distinct clustering patterns (Figure 1A). PC1 identified serum protein expression, while PC2 highlighted cellular fractions and molecular markers, such as CD103, CD161, and SLAMF7, within specific cell populations (Figure 1B). RandomForestClassifier achieved an accuracy score of 0.53, with diagnostic accuracies of 0.71 for idiopathic inflammatory myopathy, 0.64 for systemic lupus erythematosus, and 0.63 for IgG4-related disease (Figure 2A). RandomForestRegressor identified the top 30 disease-specific biomarkers, characterized by inflammatory and cytotoxic properties (Figure 2B).
Conclusion: The integration of deep immunophenotyping and serum proteomics with machine learning analysis demonstrated the potential to reveal novel biomarkers associated with inflammatory and cytotoxic characteristics. These biomarkers may provide insights into disease-specific mechanisms and support the development of refined diagnostic approaches in the future.
REFERENCES: NIL.
Principal component analysis (PCA) of CyTOF and Olink proteomics data.(A) PCA results of combined datasets of CyTOF and Olink proteomics. (B) Top 10 contributing features to PC1 and their respective weights. (C) Top 10 contributing features to PC2 and their respective weights.
Machine-learning based classification and feature selection for rheumatic and musculoskeletal disease (RMD) diagnosis. (A) Diagnostic accuracy achieved using RandomForestClassifier, illustrating the model’s performance across various RMDs. (B) Importance of top disease-specific features highlighting the contribution for RMD diagnosis.
Acknowledgements: NIL.
Disclosure of Interests: Kotaro Matsumoto: None declared, Masaru Takeshita: None declared, Katsuya Suzuki: None declared, Yuko Kaneko Chugai Pharmaceutical Co., Ltd.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (