

Background: Rheumatoid arthritis (RA) is a chronic disease affecting over 400,000 adults in the UK, causing irreversible joint damage in most patients. Methotrexate (MTX), the first-line treatment for RA, is prescribed to 60–88% of patients; however, around 40% fail to achieve satisfactory improvement. Stratified medicine offers a solution by tailoring treatment based on a patient’s biology, improving outcomes, and reducing unnecessary exposure to toxic medications. This approach depends on robust biomarkers to predict treatment response, enabling better therapeutic decisions. Metabolomic profiling has been useful in RA research, identifying lipids such as cholesterol to be associated with disease detection and treatment response, and amino acid profiles have been linked to treatment response, and disease pathogenesis.
Objectives: To identify blood-based metabolomic biomarkers of MTX response in patients with early RA.
Methods: Whole blood samples were collected from patients taking part in the Rheumatoid Arthritis Medication Study (RAMS) before starting treatment, an observational longitudinal cohort of early RA patients starting MTX for the first time in the UK. Two groups were randomly selected from RAMS based on biological sample availability forming the discovery (n=100) and validation cohort (n=189) where the samples were processed for metabolomic profile independently. Treatment response was determined after 6 months on drug and patients were classified as responders (with good and moderate responders grouped together) or non-responders using established EULAR response criteria. Two-hundred and fifty targeted metabolites were quantified in serum samples from the same patients as above using nuclear magnetic resonance (NMR)-based technology. Metabolomics data were normalised using probabilistic quotient normalization and scaled using Pareto-scaling. Partial least-square regression (PLSR) was applied to identify differentially expressed metabolites between the two groups, the input features for PLSR comprised of the metabolites along with BMI, sex, and baseline Disease activity score in 28 joints with C-reactive protein (DAS28-CRP) score.
Patient Characteristics of early RA patients participating in the study for both the discovery and validation cohorts. DAS28—CRP: Disease Activity Score in 28 joints with C-reactive protein; csDMARD: conventional synthetic disease modifying anti-rheumatic drugs; HAQ Score: Health Assessment Questionnaire score; BMI: body mass index; Disease duration measured in years; steroid usage indicates any type of steroid treatment.
| Discovery Cohort | Validation Cohort | ||||||
|---|---|---|---|---|---|---|---|
| Overall | Responders | Non-responders | Overall | Responders | Non-responders | ||
| n | 100 | 69 | 31 | 189 | 94 | 95 | |
| Sex, n (% ) | Female | 79 (79.0) | 51 (73.9) | 28 (90.3) | 125 (66.1) | 62 (66.0) | 63 (66.3) |
| Concurrent Dmards usage, n (% ) | Yes | 23 (23.0) | 14 (20.3) | 9 (29.0) | 28 (14.8) | 12 (12.8) | 16 (16.8) |
| Age, mean (SD ) | 59.9 (12.4) | 60.6 (12.3) | 58.3 (12.5) | 60.0 (13.2) | 60.0 (13.9) | 60.0 (12.6) | |
| Baseline DAS28-CRP, mean (SD ) | 4.7 (1.0) | 4.8 (1.0) | 4.4 (0.9) | 4.2 (1.5) | 4.6 (1.3) | 3.9 (1.5) | |
| HAQ Score, mean (SD ) | 1.4 (0.7) | 1.3 (0.7) | 1.5 (0.8) | 1.1 (0.7) | 1.1 (0.8) | 1.1 (0.7) | |
| BMI, mean (SD ) | 29.4 (6.5) | 28.9 (6.4) | 30.5 (6.7) | 27.8 (5.7) | 27.7 (5.4) | 27.9 (6.0) | |
| Disease Duration, median [Q1,Q3] | 0.3 [0.2,1.3] | 0.3 [0.2,1.1] | 0.4 [0.2,2.4] | 0.3 [0.1,0.7] | 0.2 [0.1,0.5] | 0.3 [0.1,1.1] | |
| Steroid usage, n (% ) | Yes | 33 (34.7) | 23 (34.8) | 10 (34.5) | 90 (55.2) | 47 (56.6) | 43 (53.8) |
The discovery cohort made up of 100 patients categorised either as responders (n=69) or non-responders (n=31) following 6-months of MTX therapy. PLSR analysis in the discovery cohort achieved a mean area under the receive-operating curve (AUC) of 0.76±0.1 during 10-fold cross validation (10CV). This motivated validation in an independent group of patients, which was made up of 189 patients with 94 responders and 95 non-responders. The aim was to have a more balanced representation to improve machine learning performance during model fitting. This resulted in AUC of 0.61±0.11, a weaker signal than discovery cohort. However, despite the weaker AUC score, both models were assessed for variable importance and similar set of features with similar magnitude, specifically creatinine and albumin (Figure 1). These two metabolites were found to be lower concentration in responders compared to non-responders. Low albumin levels are associated with increased RA disease activity, and the C-reactive protein/albumin ratio is emerging as a useful marker for assessing inflammation and disease severity.
Partial Least Square Regression biplot for both discovery and validation cohort with features with variable importance more than 1.5 highlighted in the plot.
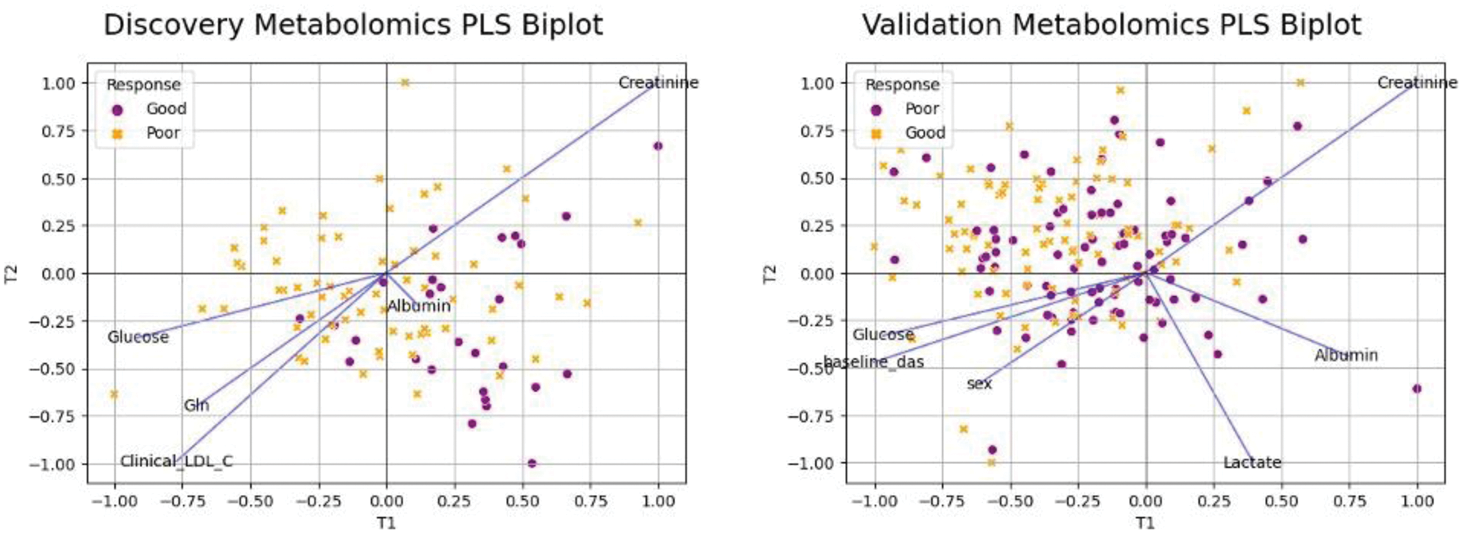
Conclusion: Our findings demonstrate that while the predictive performance of the model in the validation cohort (AUC 0.61±0.11) was weaker compared to the discovery cohort (AUC 0.76±0.1), the consistent identification of creatinine and albumin as key variables across both cohorts highlights their potential as biomarkers for methotrexate treatment response in patients with early RA. The observed lower concentrations of these metabolites in responders underscore their relevance in stratifying patients for personalised treatment approaches, contributing to improved outcomes and reduced treatment burden in RA. The weaker predictive performance in the validation cohort warrants further investigation to explore potential causes, including clinical characteristics, alternative patient outcomes, and integration with other omics data. Further studies are needed to validate these findings and assess their clinical utility.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: Chuan Fu Yap: None declared. Nisha Nair: None declared. Eryn Doe: None declared. S. Verstappen BMS and AbbVie, Kimme Hyrich: None declared. Anne Barton: None declared. Darren Plant: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (