

Background: Patients with interstitial lung disease (ILD) associated with connective tissue diseases (CTDs) suffer from higher disease-related morbidity and mortality [1]. Even though the accurate diagnosis of CTDs-ILD is essential, it often remains a challenge. This is due partially to the similarity of CTDs-ILD with other ILDs, in particular with idiopathic pulmonary fibrosis (IPF), the most severe and common ILD with different prognoses and therapies than CTDs-ILD [1]. In this regard, inflammasomes are cytoplasmic protein complexes that play a critical role in protecting the host by initiating inflammation [2]. Thus, inflammasomes have been involved in several autoimmune diseases, with the nucleotide oligomerization domain leucine-rich repeat pyrin-domain containing protein 3 (NLRP3) being the most relevant [3]. Interestingly, polymorphisms in the genes that codified the components of the inflammasomes are receiving increasing attention [3]. Accordingly, it is plausible to think that NLRP3 could have a relevant role in CTDs-ILD and may be useful in differentiating this disease from other ILDs where inflammation is not the main process involved.
Objectives: This work aimed to elucidate the role of the NLRP3 polymorphisms in the accurate diagnosis of CTDs-ILD.
Methods: Peripheral venous blood was collected from a total of 737 Caucasian patients with ILDs comprising 154 patients with CTDs-ILD, and 583 patients with ILDs without an underlying CTD (ILDs non-CTDs), of whom 242 had IPF. All patients were genotyped for NLRP3 rs4925659, rs10159239, rs10754558, rs4353135 polymorphisms by qPCR TaqMan assays.
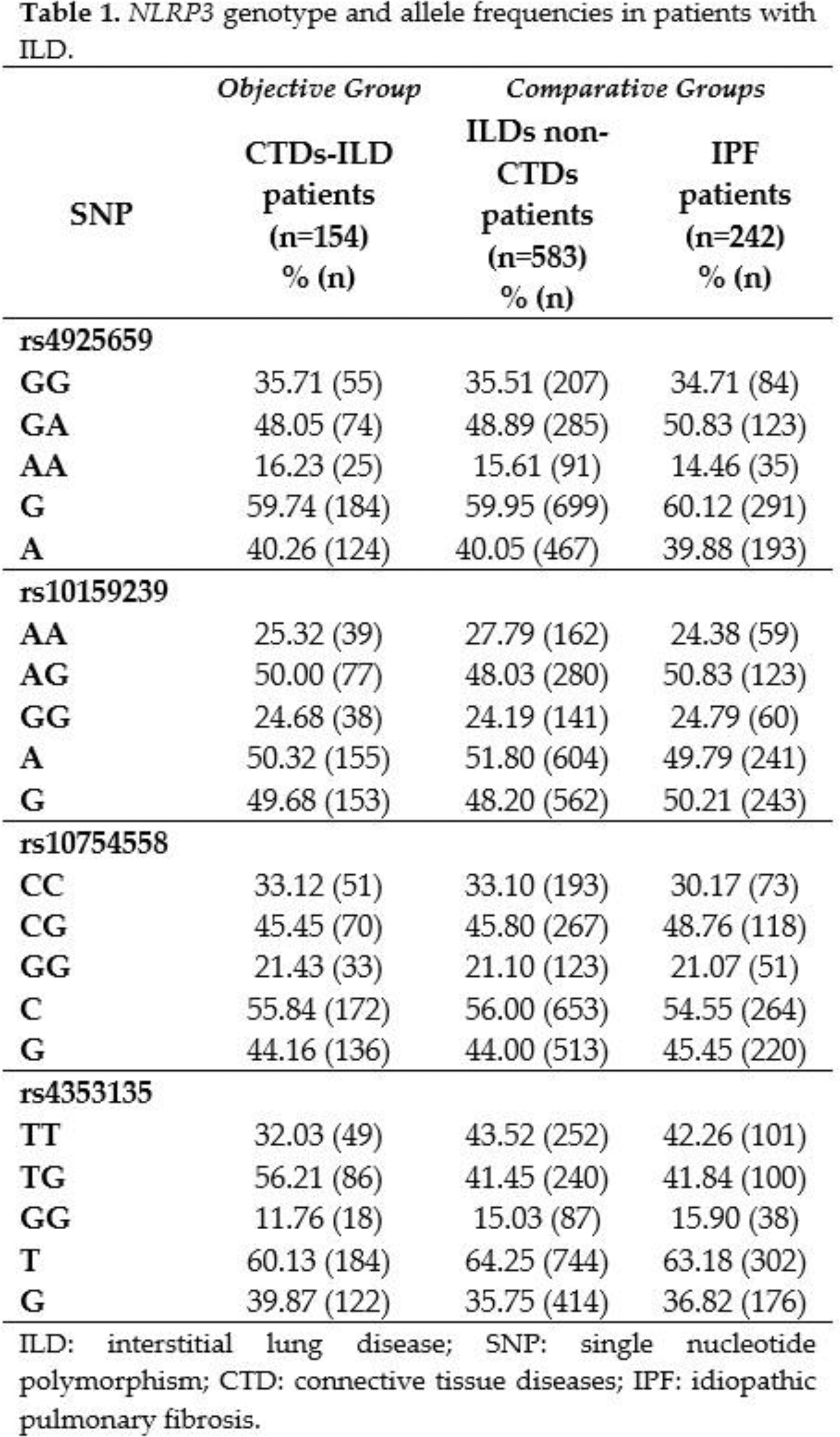
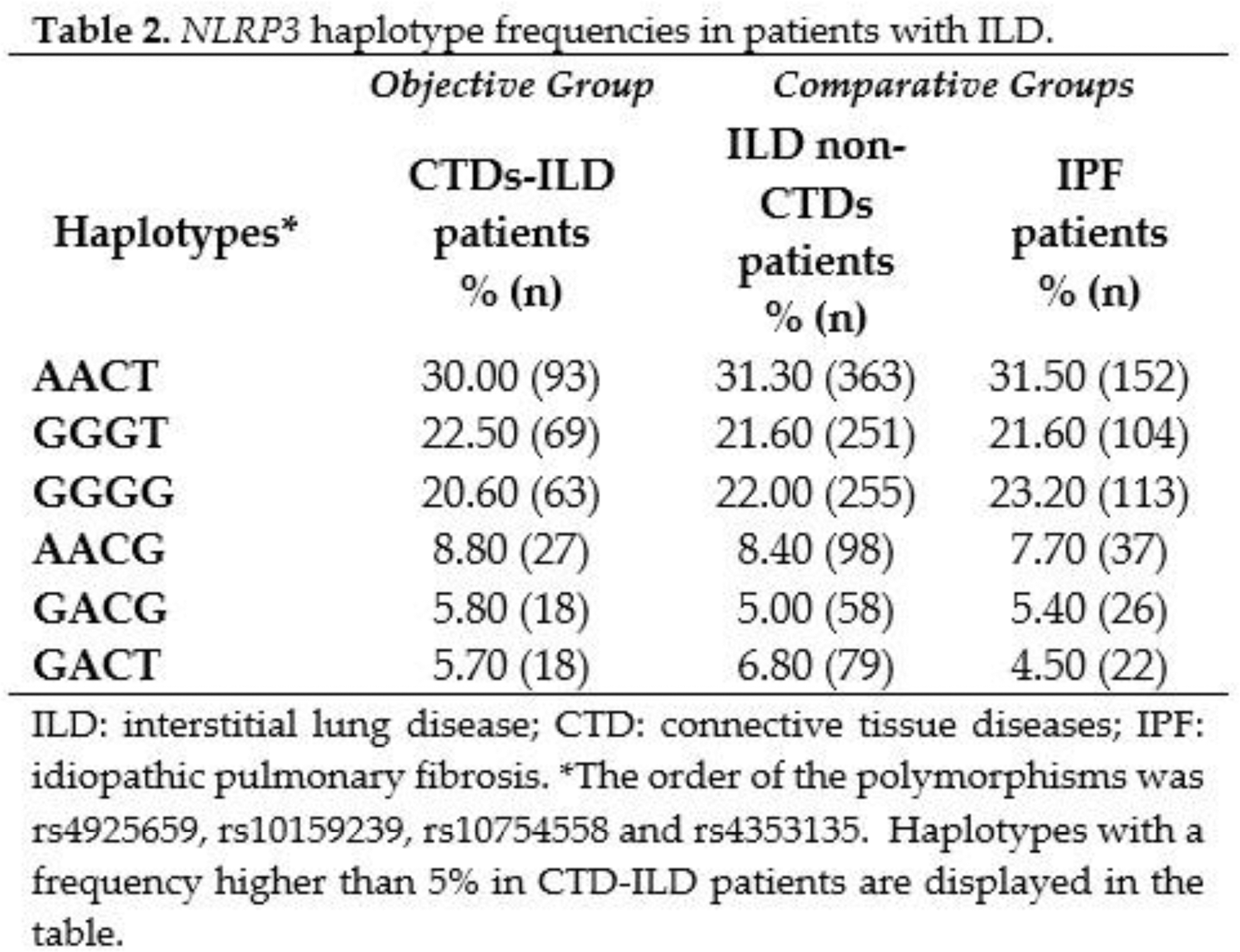
Results: The NLRP3 rs4925659, rs10159239, rs10754558, rs4353135 genotype distribution was in Hardy-Weinberg equilibrium. Genotype and allele frequencies were consistent with data from the 1000 Genomes Project for Europeans. The genotype, allele, and haplotype frequencies of NLRP3 were compared between the whole cohort of patients with CTDs-ILD and patients with ILDs without an underlying CTD, and no statistically significant differences were found between these groups (Tables 1 and 2). Specifically, we also assessed the differences in NLRP3 frequencies between patients with CTDs-ILD and those with IPF. In this regard, no statistically significant differences in the genotype, allele, and haplotype frequencies were disclosed between them (Tables 1 and 2).
Conclusion: Our findings do not support the relevant role of the polymorphisms studied in NLRP3 in the accurate diagnosis of CTDs-ILD, not being useful as biomarkers for discriminating it from other ILDs.
REFERENCES: [1] Expert Rev Clin Immunol.2018;14(1):69-82.
[2] MedComm. 2023 Oct 9;4(5):e391.
[3] J Autoimmun. 2015 Jul:61:1-8.
Acknowledgements: VP-C is supported by funds of NVAL23/02 from IDIVAL; JCB-L is a recipient of a PFIS program fellowship from the ISCIII, co-funded by the European Social Fund (‘Investing in your future’), (FI22/00020); and RL-M is a recipient of a Miguel Servet type II program fellowship from the ISCIII, co-funded by ESF (“Investing in your future”) (CPII21/00004).
Disclosure of Interests: Verónica Pulito-Cueto Boehringer Ingelheim, Joao Carlos Batista-Liz: None declared. Belén Atienza-Mateo: None declared. David Iturbe Fernández: None declared. Victor Manuel Mora-Cuesta: None declared. María Sebastián Mora-Gil: None declared. Carolina Aguirre Portilla: None declared. José M. Cifrián-Martínez: None declared. Ricardo Blanco had consultation fees/participation in companysponsored speaker´s bureau from Abbvie, Pfizer, Roche, Bristol-Myers, Lilly, Janssen, and MSD, has received grants/research supports from Abbvie, MSD, and Roche, Raquel López-Mejías: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (