

Background: Fatigue is highly prevalent among patients with psoriatic arthritis (PsA) significantly affecting patients’ quality of life [1]. Existing evidence suggest an association between fatigue and female sex, inflammation, pain, etc [2]. However, the underlying molecular mechanisms of fatigue remain poorly understood, likely due to the complex and multifactorial nature of fatigue [3].
Objectives: The primary aim of the study was to uncover the underlying genetic components associated with severe fatigue in PsA patients using data retrieved from single cell RNA sequencing (scRNAseq).
Methods: Patients were included from the Parker Institute’s PsA patient cohort [4] with scRNAseq data available from 70 PsA patients and 10 sex and age-matched healthy controls (HCs). Peripheral blood mononuclear cells (PBMCs) were processed with scRNAseq retrieving ~9,950 cells per sample, which were sequenced to a depth of 50,000 reads per cells (paired-end sequencing, 2x150bp). PsA patients were stratified based on Visual Analogue Scale (VAS) fatigue score (0-100mm), i.e., none-to-mild (N2M) fatigue (VAS fatigue ≤20mm) and severe fatigue (VAS fatigue >80mm). PsA patients with VAS fatigue >20 mm and ≤80 mm were excluded from the study. An analysis of differentially expressed genes (DEGs) were used to compare 1) PsA patients with severe fatigue vs N2M fatigue, and 2) PsA patients with severe fatigue vs HCs. Identified DEGs were included in gene set enrichment analyses (GSEA) for further examination of the impact of influential genes on biological pathways (gene ontology; BP) in association to fatigue. Ancillary post-hoc analyses were performed implementing a sex-specific normalization based on a standardized z-score for male and female.
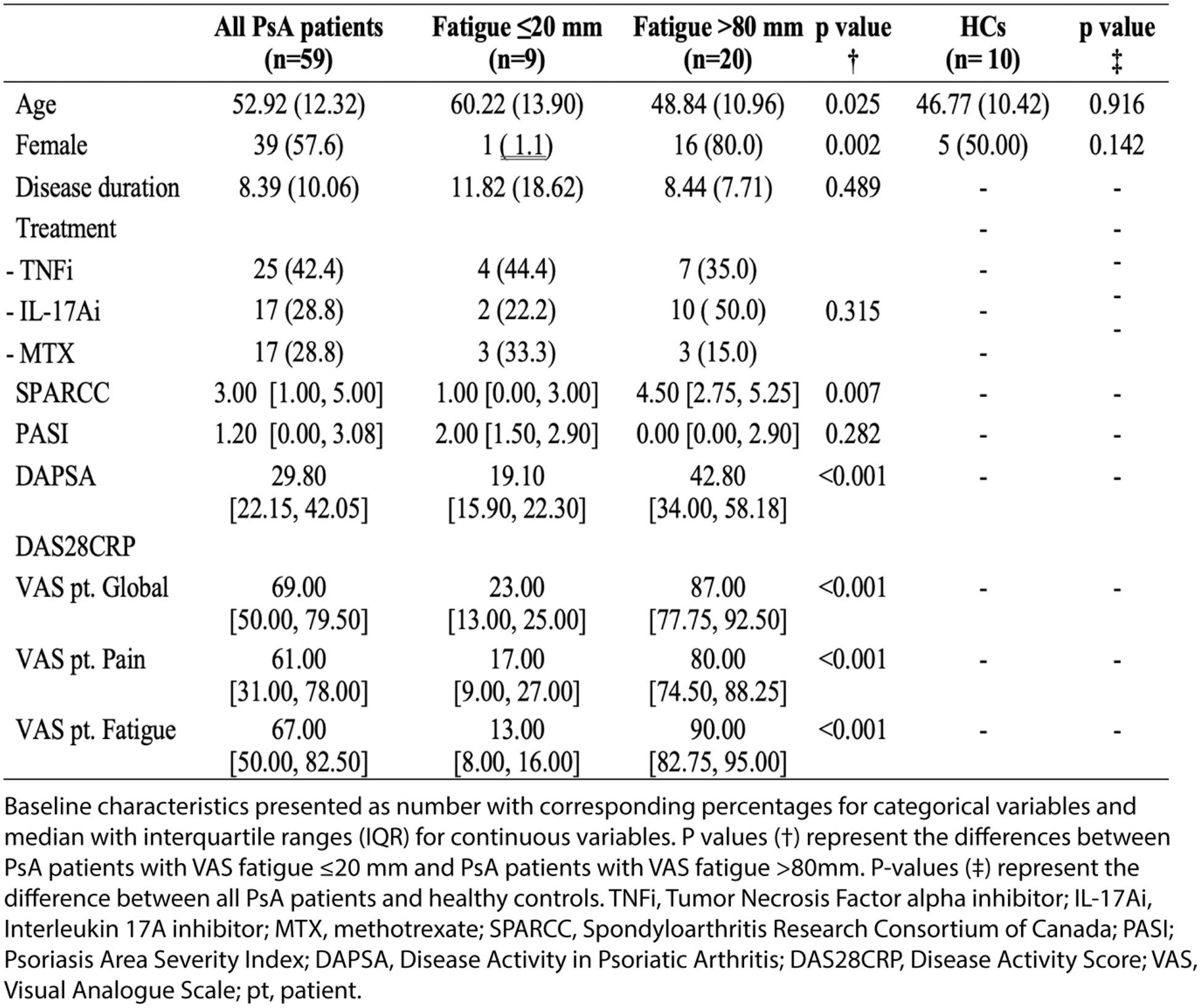
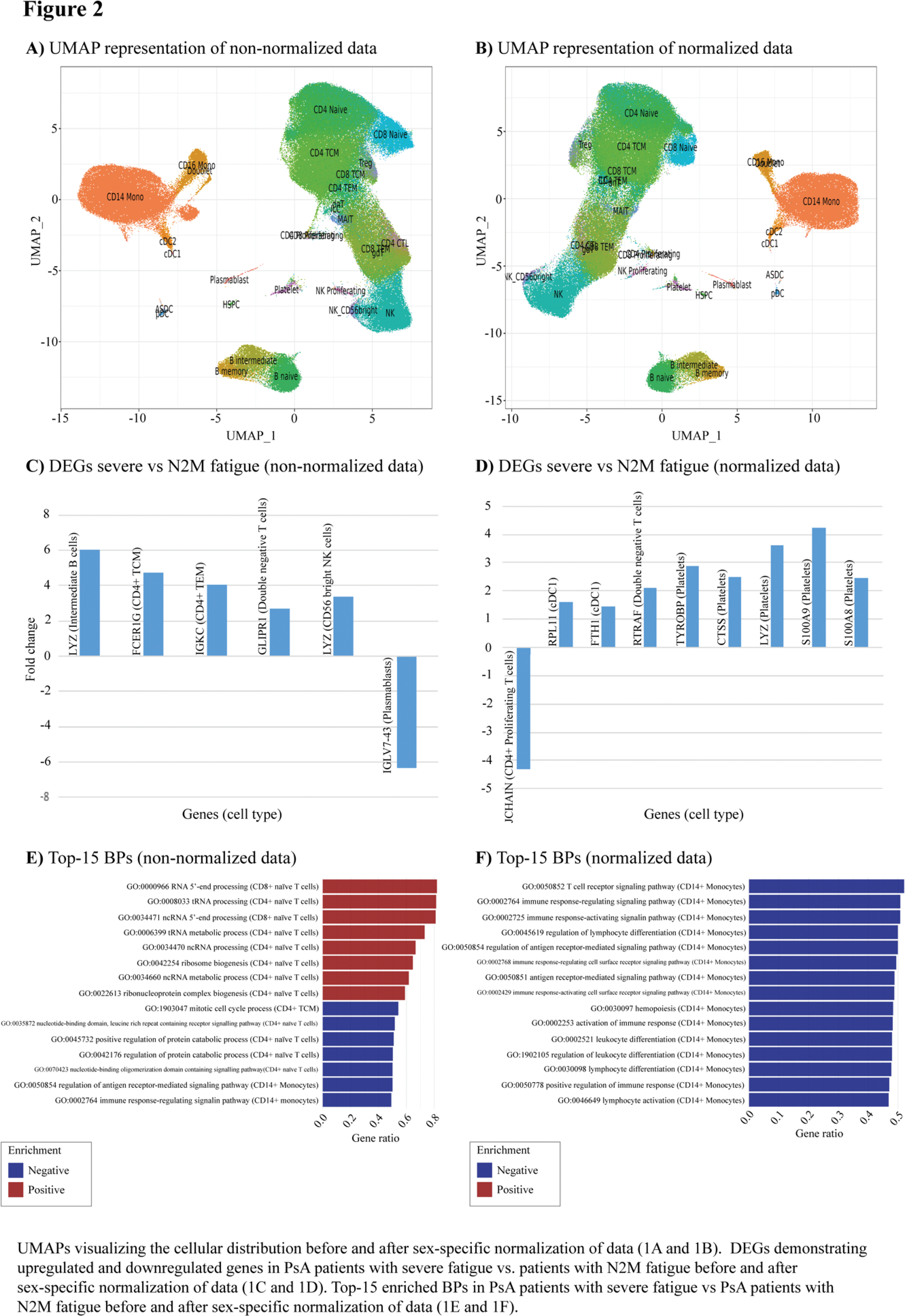
Results: ScRNAseq data from 29 PsA patients were included for the analysis, including 9 patients with N2M fatigue and 20 patients with severe fatigue (Table 1). PsA patients suffering from severe fatigue were predominately female (80.0%) and younger (48.8 years [SD 11.0]) compared to patients with N2M fatigue (11.1%, p-value 0.002, and 60.2 years [SD 13.9], p-value 0.025, respectively), p-values <0.05 were considered statistically significant. Further, PsA patients with severe fatigue had statistically significant higher SPARCC score, DAPSA and DAS28CRP disease activity, VAS patient global and pain. Post-hoc analysis were performed to account for difference in sex. No significant differences in cell populations were found in the analysis of non-normalized vs normalized data (Figure 1A-B). The comparison of non-normalized data from patients with severe vs N2M fatigue revealed upregulated DEGs included FCR1G, GLIPR1 , and LYZ , and down-regulated IGLC7-43 in several immune cell subtypes (Figure 1C). While the analysis of normalized data revealed upregulated DEGs, including CTSS, LYZ, RTRAF, TYROBP, S100A8 and S100A9 , and down-regulated CXCL9 in various immune cell and platelets (Figure 1D). The comparison of PsA patients with severe fatigue vs HCs demonstrated DEGs associated with PsA-associated inflammation, including upregulation of STEAP3 . GSEA incorporating non-normalized data demonstrated the influence of CD4+ and CD8+ T cells, which in PsA patients with severe fatigue exhibited significant positive enrichment of pathway associated with the biological pathways; maturation of non-coding and transfer RNA, and processes related to synthesis of proteins and ribonucleosome complexes, compared to patients with N2M fatigue (Figure 2E). Twelve out of top-30 enriched pathways, i.e., the thirty BPs with the highest gene ratio, were shared independent of the utilization of non-normalized or normalized data. All shared BPs were downregulated in CD14+ monocytes and related to activation and regulation of the immune response but exhibited higher gene ratio using normalized data (Figure 2F). Two out of the top-30 enriched BPs were upregulated in PsA patients with severe fatigue compared to HCs. Interestingly, both pathways were involved in rhythmic processing, including circadian rhythm (GO:0048511, GO:0007623, enrichment score>0.4).
Conclusion: This was an exploratory study investigating underlying mechanisms of fatigue in PsA. Several upregulated DEGs, including LYZ, CTSS, S100A8 and S100A9 , have been associated with neuroinflammation. Additionally, STEAP3 has been identified as a contributor to neuroinflammation in autoimmune disorders mediated by T helper cells type 17 [5], suggesting a possible link to experienced fatigue of PsA patients. Notably, LYZ, CTSS, S100A8 and S100A9 were all upregulated in platelets. Platelets are rarely discussed in relation to the inflammatory activity of PsA. However, their established role in interacting with immune cells during inflammation suggests they may warrant further investigation as potential contributors to fatigue experienced in PsA. Study results should be interpreted in the context of observed differences in sex and disease activity measures, as these factors are likely to influence gene expression. Future research may benefit from focusing on the mechanisms underlying fatigue within specific patient subgroups further stratified by sex and disease activity. However, given that the mechanisms driving fatigue likely vary depending on patient and disease characteristics, tailored strategies may be necessary for effectively managing fatigue in PsA patients.
REFERENCES: [1] Gossec, et al. J Rheumatol. 2022 Nov;49(11):1221-1228.
[2] Skougaard, et al. J Rheumatol. 2020 Apr;47(4):548-552.
[3] Gudu, et al. Joint Bone Spine. 2016 Jul;83(4):439-43.
[4] Højgaard, et al. BMJ Open. 2016 Apr 15;6(4).
[5] Horstman, et al. J Neuroinflammation. 2010 Feb 3;7:10.
Patient characteristics.
Acknowledgements: The authors wish to acknowledge the patients for their contribution to the study.
Disclosure of Interests: Zara Rebecca Stisen: None declared. Melanie Randahl Nielsen: None declared. Sisse B. Ditlev: None declared. Leon Eyrich Jessen: None declared. Marie Skougaard Janssen-Cilag, Eli Lilly, Pfizer.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (