

Background: Systemic sclerosis (SSc) is a systemic autoimmune disease characterized by endothelial dysfunction, vasculopathy, immune dysregulation, and subsequent fibrosis of the skin and visceral organs. Despite a broad range of treatment options, the disease still leads to progressive disability, diminished quality of life, and mortality. An analysis of the results of relevant global studies about genetic mutations, spectrum of antibodies and cytokines in patients with SSc could identify commonalities, differences and gaps in existing knowledge and finally improve understanding of the pathogenesis of this disease.
Objectives: to assess the susceptibility genes, antibodies and cytokines involved in SSc in Kazakh cohort of patients.
Methods: Possible genetic mutations, antibodies and cytokines, involved in autoimmune aggression in SSc were identified through a systematic review of relevant literature from biomedical databases, as well as by comparing with genetic panels offered by leading manufacturers. A comprehensive literature search was conducted using PubMed and Google Scholar databases with keywords such as “SSc”, “genetic mutations”, “sequencing, “NGS,” and “exome”. Priority was assigned to peer-reviewed research articles, systematic reviews, and meta-analyses. Additionally, the Ion AmpliSeq™ Designer gene repository was utilized to identify disease-associated genes. A total of 30 Kazakh individuals with diffuse form of SSc were recruited and examined for disease activity, skin and internal organs impairment using EScSG and Rodnan skin score (RSS). Interleukin (IL)-1β, 4, 6, TNF-α were evaluated by ELISA. Antinuclear factor (ANF) was estimated by HEp2 cells. Antibodies to SCL-70, U1-snRNP, CENP-B, SS-A/Ro52, SS-A/Ro60, Sm/RND, Sm, SS-B, Rib-P0, nucleosomes, Jo-1 were determined by immunoblotting. This study employed a custom AmpliSeq panel on an Ion Proton sequencer to investigate 120 genes potentially associated with SSc. Control samples for sequencing were obtained from 18 individuals. Variants were analyzed using Ion Reporter software.
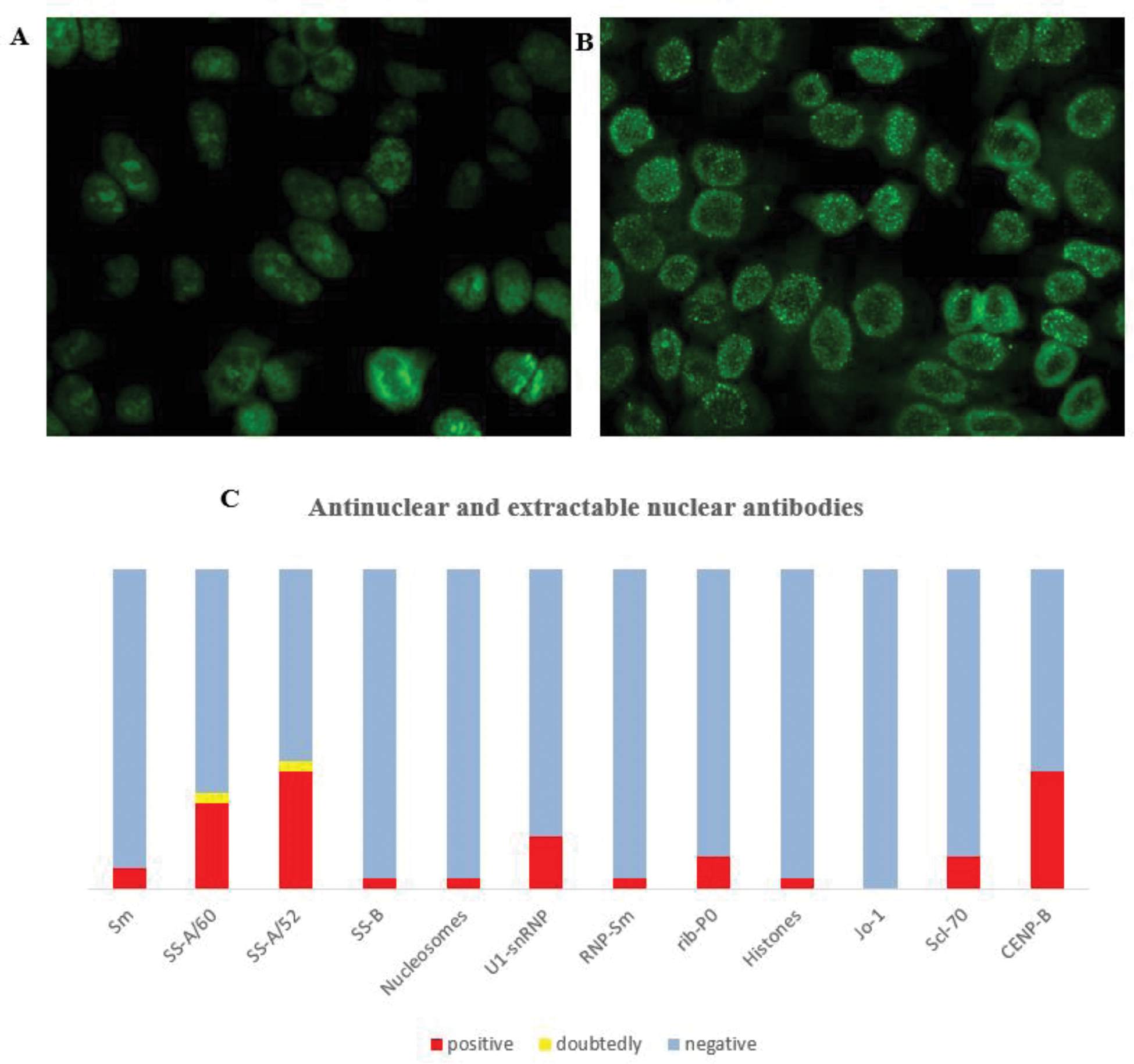
Results: Between 1965 and 2024, a total of 4,367 publications were identified that discussed the subject matter mentioned above, only 223 of these were match the criteria and included in this analysis. Antinuclear antibodies (ANAs) were detected in 95% of patients anti-SCL-70, anti-topoisomerase I, anti-RNA polymerase III, anticentromere antibodies were found in 223 research 12 studies revealed an association of active SSc with an increase of TGFβ, IL-1, IL-6, IL-33, IL-4, IL-13, IL-10, MCP-1, IFN-γ, TNF-α, IL-1α, IL-2 and CXCL10 cytokines. A total of 35 research identified the associations SSc with genetic polymorphisms in ANKS1A, ATG5, C6orf10, CD247, GRB10, ITGAM, IRF5, JAZF1, NOTCH4, PSORS1C2, PXK, RHOB, SOX5, STAT4, TNPO3, CLEC16A. Based on the results of the current review, a preliminary list of genes, autoantibodies, and cytokines associated with the development of SSc was prepared. Analysis of data obtained from 30 Kazakhstani SSc patients (RSS 16.2±7.73; EScSG 3.33±2.19) revealed antibodies to CENP-B, SS-A/Ro60, SS-A/Ro52, SS-B, Scl70, U1-snRNP, Sm, Rib-P0, nucleosomes, histones (Figure 1). Cytokines level were about normal.
In the group of patients with SSc, a change in the LY96 gene is noted, the detected variant chr8:74922341 CT/C is found in 11 patients. The variant is classified as a variant of unknown clinical significance. The gene encodes a protein that binds to toll-like receptor 4 on the cell surface and provides sensitivity to lipopolysaccharide (LPS), thereby providing a link between the receptor and LPS signalling. In addition, changes are noted in the IRAK1 gene. The chrX:153278833 GCCCG/GCC variant is found in 2 patients and one participant in the control group, and the chrX:153278833 GCC/GCCG variant is found in 3 patients of the study group and does not occur in the control group. Both variants are classified as variants of unknown clinical significance. Four patients have different variants in the AIRE gene (chr21:45708278 G/C, chr21:45711068 TC/T, chr21:45713024 A/G, chr21:45711025 C/G), which were not found in the control group. The AIRE gene encodes a transcriptional regulator that forms multimeric nuclear protein complexes in superenhancer regions of chromatin in thymic medullary epithelial cells (mTEC), where it mediates the expression of tissue-specific antigens and promotes immunological self-tolerance. The chr21:45711068 TC/T variant is classified as likely pathogenic, the other three variants as of unknown clinical significance.
Conclusion: The immunogenetic panel developed through this analysis offers a targeted approach to unraveling the genetic basis of SSc for early diagnosis, tailored treatment strategies, and a more profound comprehension of the molecular mechanisms underpinning this autoimmune disorder. Remarkably, this is the first genetic study of SSc patients conducted in Kazakhstan. It has the potential to contribute to the expansion of the current genetic panel and the discovery of novel insights into the pathogenesis of the disease and underscores the importance of considering the full spectrum of genetic variation in SSc pathogenesis.
Antinuclear and extractable nuclear antibodies: (A) Antinuclear factor on HEp2 cells estimated by indirect immunofluorescence, titre 1:640, nucleolar type of glow; antibodies to CENP-B; (B) Antinuclear factor, titre 1:1280, сentromeric type of luminescence; antibodies to SS-A/60, SS-A/52 and CENP-B; (C) Spectre of antibodies (persentage of ANA/ENA) revealed in SSc patients.
REFERENCES: NIL.
Acknowledgements: This abstract was undertaken as part of research funded by the Ministry of Science and Higher Education of the Republic of Kazakhstan (grant number АP19576783).
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (