

Background: Behçet’s Disease (BD) is a chronic, systemic inflammatory disorder. While primary triad consists of oral and genital aphthous ulcers, and uveitis, the vascular, neurological, and gastrointestinal systems may also be involved during the course. The severity, course, and management principles vary according to the subphenotypes formed by different combinations of involved organs and patients’ age and sex. Clarifying the pathogenic features of the subphenotypes may enable us to use more effective therapeutic approaches depending on the organs involved.
Objectives: This study aimed to investigate the differing roles of innate and adaptive immune responses in three main phenotypes of BD (mucocutaneous, ocular, and vascular), understand their effects on immunopathogenesis, and provide data to optimize the use of immune-targeting agents or avoid inappropriate therapies based on the phenotypes.
Methods: The serum samples were collected during the active and post-treatment remission phase of BD patients and classified according to phenotypes. Patients with overlapping organ involvements or gastrointestinal/neurologic involvements were excluded. Serum concentrations of the pro- and anti-inflammatory cytokines were measured using sandwich ELISA. Transcriptome analyses were performed using RNA sequencing of the peripheral blood mononuclear cells (PBMCs). RNA libraries were prepared using the Illumina Ribo-Zero Library Prep Kit. RNA quality was assessed using RNA IQ Qubit. Sequencing was performed on Illumina NovaSeq6000 platform (PE150) with S4 chips. The raw RNA-seq data underwent quality control using FastQC (v0.11.9) and were trimmed with Trimmomatic (v0.39) to remove adapters, indices, and low-quality reads. The cleaned sequences were aligned to the human reference genome (hg38, GRCh38) using HISAT2 (v2.2.1), and read counts were generated with HTSeq (v2.0.8) using the GENCODE hg38 annotation (v.110 gtf). Differentially expressed gene analysis (DEG) were performed using the DESeq2 package (v1.38.3) within the R Bioconductor platform (v4.2.3), with additional normalization and analysis using the NOISeq package (v2.42.0) for conditions lacking biological replicates. The roles of innate and adaptive immune systems were analyzed through DEG results among the groups.
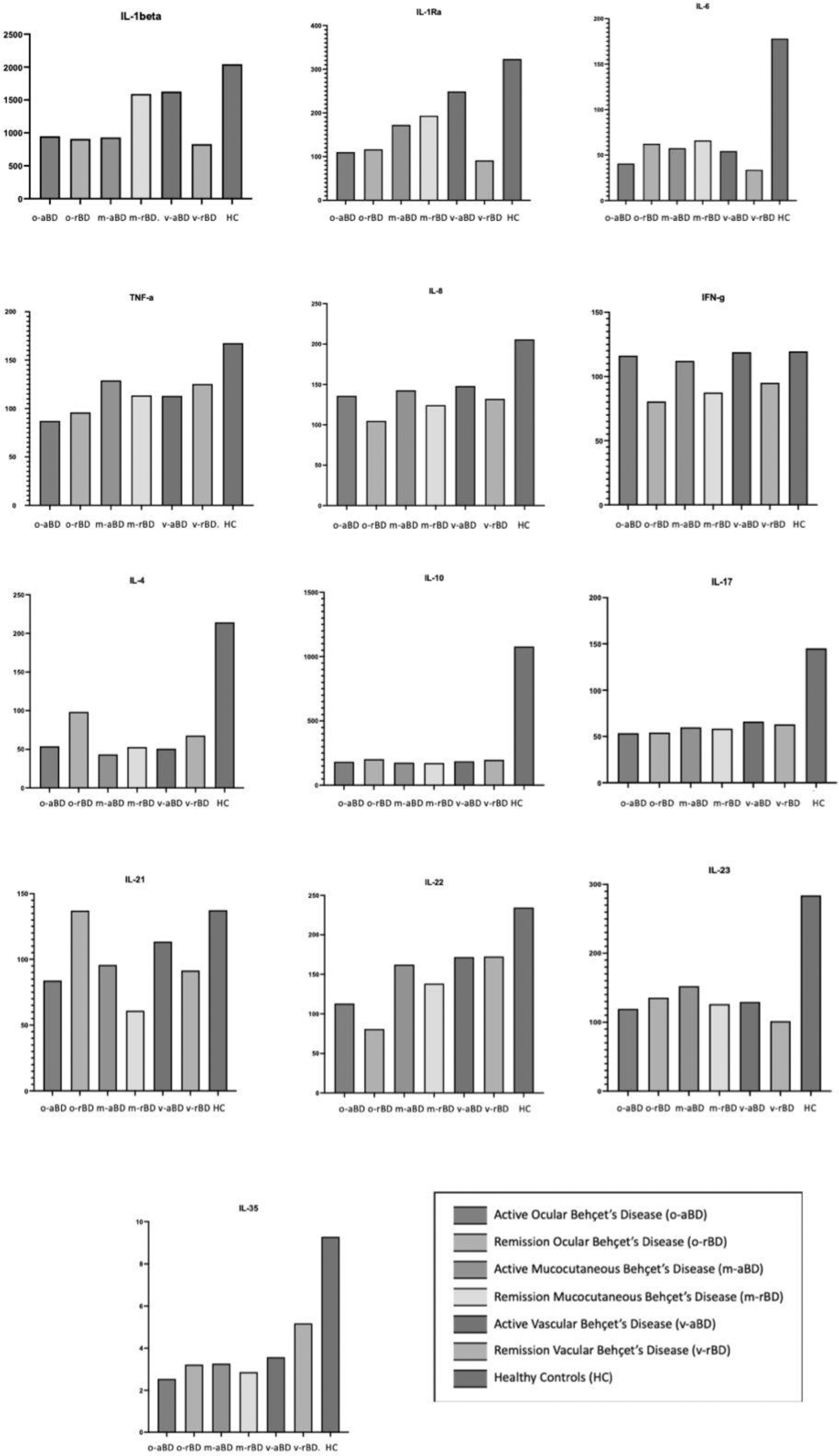
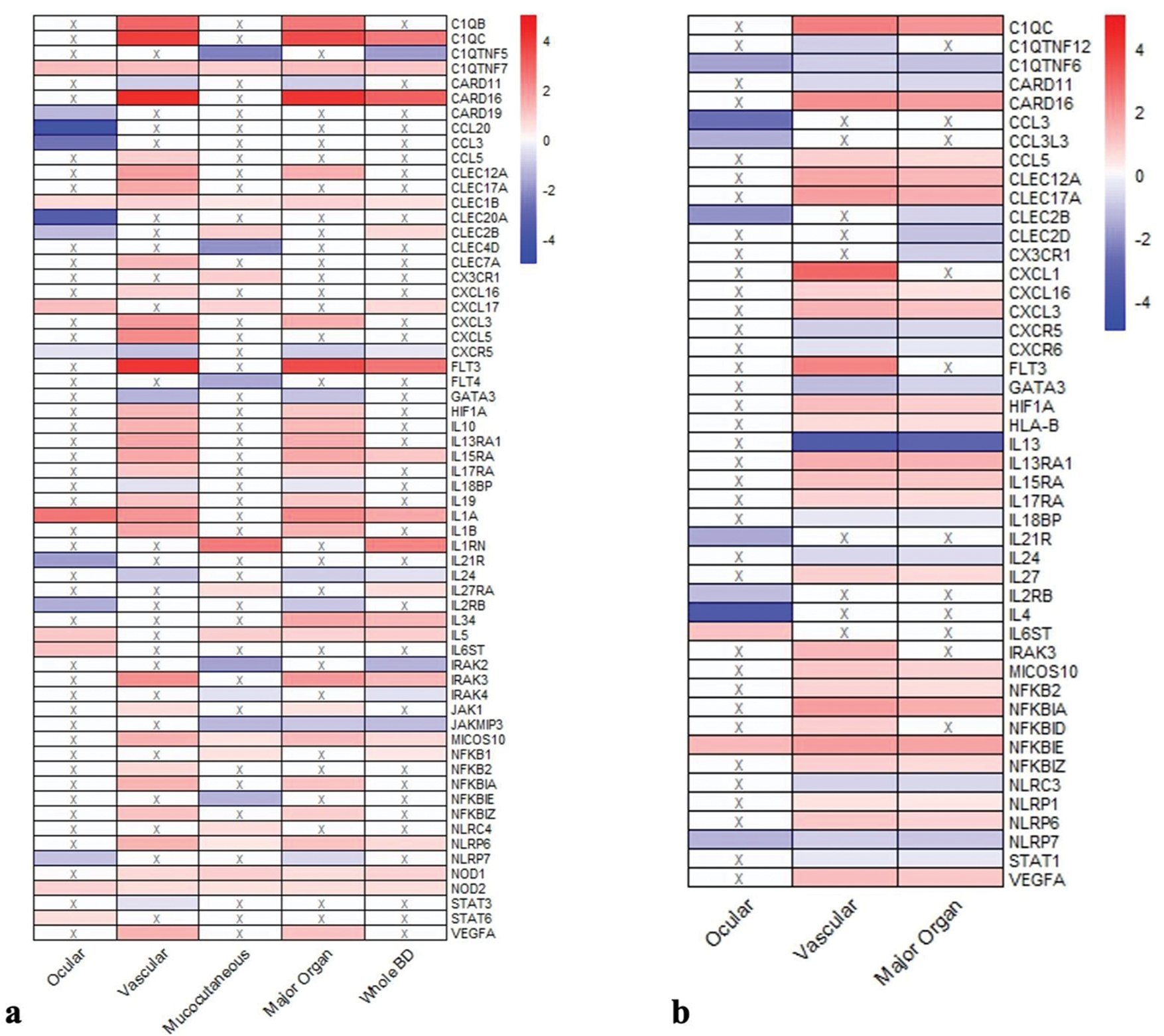
Results: The ELISA results are given in detail in Figure 1. The analysis was performed in 41 BD patients with active disease (aBD) (M/F:9/32, median age 29[18–46]), 35 patients in remission (rBD) (M/F:9/26, median age 29[18–46]), and 9 healthy controls (HC) (M/F:3/6, median age 28[23–32]). The aBD group included 19 patients with mucocutaneous involvement (m-aBD), 11 with ocular involvement (o-aBD), and 11 with vascular involvement (v-aBD). Serum IFN-gamma levels were significantly higher in the aBD group compared to the rBD group (116vs.92 pg/ml, p=0.022). Serum IL-35 levels were significantly higher in the HC group than in the aBD and rBD groups(p=0.05). IL-17-related cytokines were lower in the ocular aBD group. These cytokines increased after treatment in ocular but decreased in mucocutaneous and vascular aBD groups. Transcriptome analyses were conducted with the samples of aBD patients, including 29 with m-aBD, 10 v-aBD, and 4 o-aBD, along with 15 HC. Significant differences were observed in the expression profiles of genes involved in innate immunity, inflammasome activation, adaptive immunity, and regulating these inflammatory processes, which are summarized in Figure 2. Like ELISA findings, IL-17-related cytokine expressions were reduced in the o-aBD group. The certain genes related to angiogenesis and hematopoiesis (VEGF, HIF1A, and FLT3) were expressed 4–13 times more in the v-aBD group.
Conclusion: This study supports the roles of both innate immunity and Th1-dominant adaptive immune responses in BD pathogenesis. Reduced IL-17 and Th17-related immune responses in ocular BD may explain the lack of favorable response with IL-17 blockade in patients with uveitis. The expression levels of the genes involved in innate and adaptive immune responses differ across the BD phenotypes. Suppression of Th17-related pathways and the activation of innate immune mechanisms play a key role in determining phenotypic diversity. The vascular BD group is distinguished by significant activity in angiogenesis, hematopoiesis-related genes, and other innate immunity/inflammasome-related genes not affecting the mucocutaneous group. These findings expand the current knowledge of BD immunopathogenesis and offer critical insights for developing more precise management algorithms.
The median cytokine serum levels in three phenotypic subgroups as ocular Behçet’s Disease (BD)(o-BD), mucocutaneous (m-BD), and vascular (v-BD) in active and remission state and Healthy Controls.
Gene expression profiles in active BD patients. (a)A comparison of the gene expression profiles of ocular, vascular, mucocutaneous, major organ involvement (ocular+vascular), and all BD patients to healthy controls and (b) comparison of the gene expression profiles of ocular, vascular, and major organ BD subgroups to mucocutaneous BD patients. The heatmap colors represent the magnitude of gene expression changes as log2 fold changes: red indicates upregulation, with deeper red reflecting stronger upregulation, while blue indicates downregulation, with deeper blue reflecting stronger downregulation (An “X” in a box signifies no significant fold change for that gene). Log2 fold changes can be interpreted directly in terms of fold differences:for example, a log2 value of 2 corresponds to a 4-fold change, while a value of 3 corresponds to an 8-fold change. This visualization highlights the distinct gene expression patterns across different clinical subtypes of BD.
REFERENCES: NIL.
Acknowledgements: This study was supported by the Research Universities Support Program (ADEP) of the Marmara University Scientific Research Projects (BAP) under project number ADEP-10865.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (