

Background: Osteoarthritis (OA) is a prevalent condition among people of advanced ages and presents with pain, stiffness, and in severe cases disability. Currently, there are no approved disease-modifying drugs for OA despite advancement in our understanding of OA disease biology. This translational failure is attributed to heterogeneity within OA patients, where patients present with variable disease progression and severity.
Objectives: To characterise peripheral immunome dysregulations accompanying joint inflammation in knee OA, and to distil immune signatures for patient classification into different OA subtypes. To construct a immune cytomic-based classification tool for identification of patients with increased disease severity.
Methods: We employed mass cytometry time-of-flight (CyTOF) to investigate peripheral blood mononuclear cells (PBMCs) obtained from patients with knee OA and healthy controls. 57 patients with knee OA and 33 age, sex, and ethnicity-matched HCs were enrolled in this study. An additional 40 patients with knee OA were enrolled for our validation study. Two CyTOF marker panels that cover major immune cell lineages were designed to interrogate these samples to capture broad immune lineage changes. Resulting single-cell proteomic CyTOF data was analysed using statistical and machine learning algorithms to extract unique peripheral immune signatures for patient classification into different disease subtypes. Machine-learning models were constructed using these peripheral immune signatures to classify OA patients of our discovery and validation sets.
Results: Using a combination of statistical and machine-learning algorithms, we derived 10 immune clusters that, when used in K-means clustering, identified three OA subtypes with distinct clinical phenotypes and corresponding peripheral immune signatures. We observed an inflammatory subtype (Group 3) where patients present clinically with greater pain and stiffness, and poorer function. Patients with this inflammatory subtype have a peripheral immune signature characterised by increased monocyte and effector T/T-like immune subsets. Another OA subtype (Group 1) with an immune-modulated profile was also identified where patients report less pain and stiffness. The peripheral immune signature of this group of patients suggests increased immune regulation as seen in the relative increase in Treg and exhausted Th subsets. Group 2 patients exhibited intermediate symptoms with where their peripheral immunome are relatively increased in NK subsets suggesting an innate immune character. Machine-learning models built using frequencies of the 10 identified immune clusters were efficacious in classifying patients, with inflammatory subtype and worse symptoms, of our discovery and validation cohorts from other OA patients.
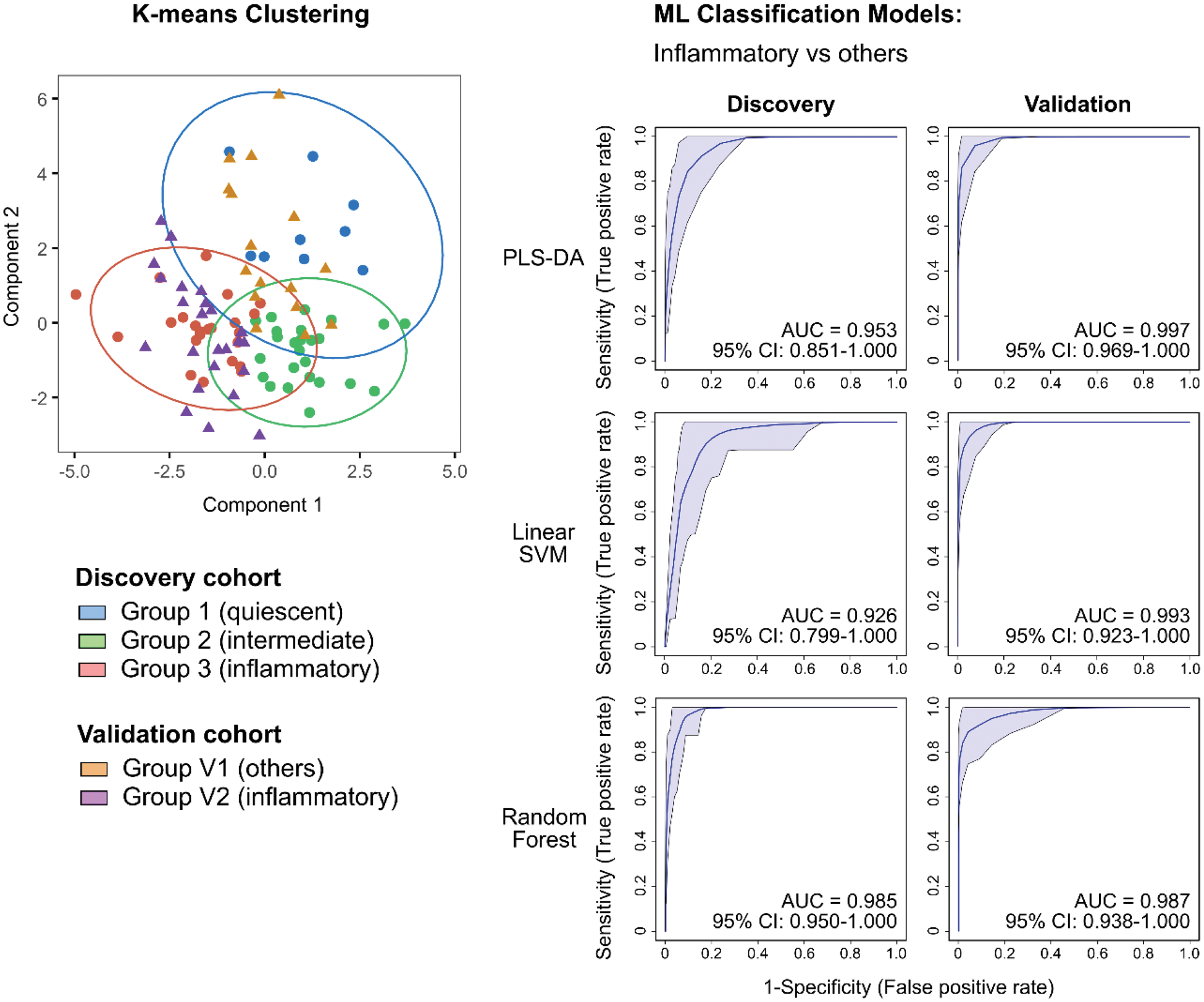
Conclusion: In conclusion, we identified a set of 10 peripheral-blood immune clusters, using a combination of statistical and machine-learning approaches in the analysis of high-dimensional cytomic data, that classified patients into three subtypes with distinct immunological differences. Findings from our validated study have clinical applications as a promising precision medicine tool in the comprehensive management of OA where pharmacotherapeutic decisions can be made based on a patient’s peripheral immunophenotype such as the identification of patients with inflammatory subtype for early immunomodulation to arrest disease progression. Furthermore, our identification of these clinical subtypes with accompanying biologically relevant immune differences provide further insights in the underlying disease mechanisms driving heterogeneity in OA patients.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (