

Background: Dermatomyositis (DM) is a chronic inflammatory myopathy characterized by symmetric proximal muscle involvement and distinctive skin lesions, including Gottron’s papules and heliotrope rash. Patients with DM carry an increased risk of developing malignancies compared with age- and sex-matched controls, having important implications for disease management and prognosis [1]. The malignancy risk in DM is significantly higher, among others, for breast carcinoma, colorectal, pancreatic adenocarcinoma, leukemia, and lung neoplasms [2]. The molecular mechanisms are not well known, and possible biomarkers of malignancy in DM could give an earlier diagnosis and better prognosis for these patients. The study identified key HGs overexpressed in DM-associated cancers. We try to bring forward a comprehension of the molecular signatures joining DM and cancer and discuss the feasibility of applying them as biomarkers for malignancy surveillance.
Objectives: The aim of the present study was to identify hub genes related to five different types of cancers: breast carcinoma, colon cancer, pancreatic adenocarcinoma, leukemia, and lung neoplasms.
Methods: The GSE1551 dataset was retrieved from the Gene Expression Omnibus platform. Gene Set Enrichment Analysis was performed to find overrepresented genes across the five malignancy types. The GSEA program was used to identify the enriched gene sets for breast carcinoma, leukemia, colon cancer, lung neoplasms, and pancreatic adenocarcinoma. Protein-protein interactions (PPIs) were analyzed using the Cytoscape platform. Overlapped HGs were identified through five algorithms: Degree, DMNC, Eccentricity, Radiality, and Stress, using the Cytohubba plugin. ROC curves showing mean fluorescence intensity values (MFI) for DM patients versus HC were constructed.
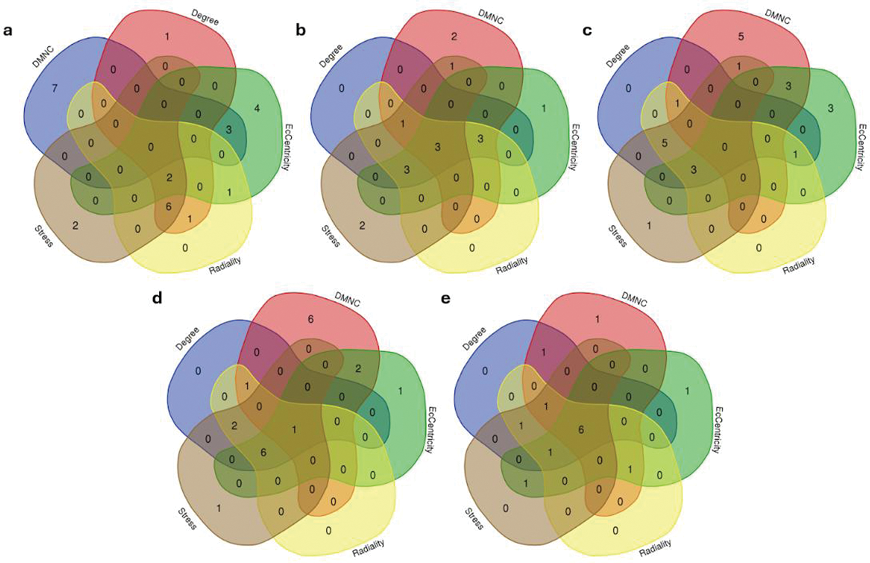
Results: A total of 13 patients with DM were compared to 10 healthy controls. Figure 1 illustrates the overlap of hub genes (HGs) identified using five different algorithms across five cancer types. Key HGs PTEN and ATM were identified for breast carcinoma; PIK3CA , BRCA2 , and ATM for leukemia; BRCA2 for lung neoplasm; MLH1 , PALB2 , and CHEK2 for colon cancer; and MLH1 , EPCAM , ATM , KRAS , SMAD4 , and TP53 for pancreatic adenocarcinoma. Interestingly, PTEN , BRCA2 , CHEK2 , EPCAM, KRAS, and TP53 genes are highly expressed in the DM patients when compared with the controls as shown in Figure 2.
Overlapping hub genes identified in dermatomyositis across five cancer types. Venn diagrams illustrate the shared hub genes (HGs) among five malignancies commonly associated with dermatomyositis: (a) Breast carcinoma, (b) Leukemia, (c) Lung neoplasm, (d) Colorectal cancer, and (e) Pancreatic adenocarcinoma. The diagrams were generated using five different algorithms (Degree, DMNC, Eccentricity, Radiality, and Stress) via the CytoHubba plugin in Cytoscape, highlighting the most interconnected and critical genes across these malignancies.
Diagnostic performance of key hub genes in dermatomyositis across multiple cancer types. The figure represents the expression levels, as determined by MFI values, of key hub genes including PTEN , ATM , PIK3CA , BRCA2 , MLH1 , PALB2 , CHEK2 , EPCAM , KRAS , SMAD4 , and TP53 , between DM patients and healthy controls. The left panels show MFI of each hub gene, while the right panels illustrate the ROC curve for the diagnostic performance of each gene in distinguishing DM patients from healthy controls. The ROC curves were plotted, and the AUCs were computed along with 95% CIs and p-values, which give indications on the power of discrimination for each gene. Statistical significance *p < 0.05, **p < 0.01 is represented here, which may be useful for these genes as biomarkers of cancer risk in DM patients.
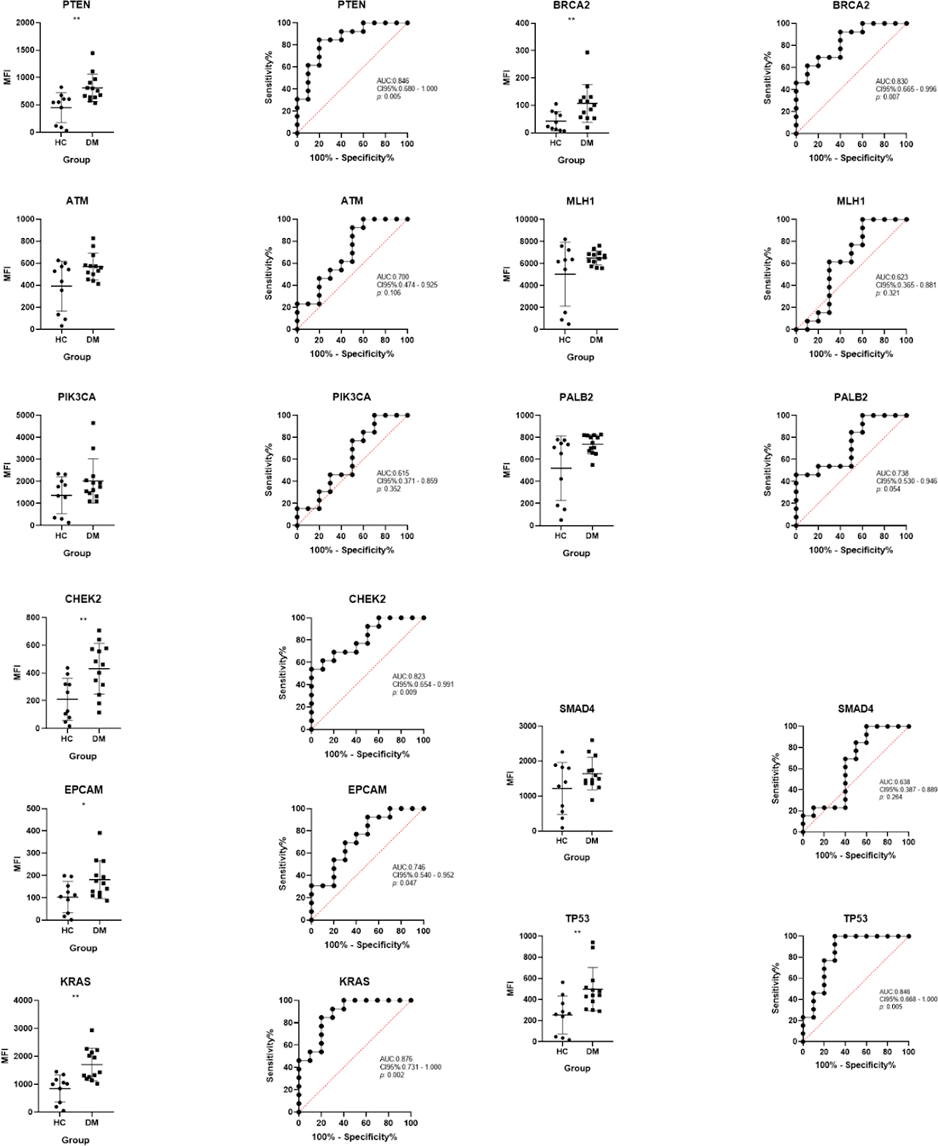
Conclusion: The present study identified several key hub genes overexpressed in DM patients with associations to five different kinds of malignancies. Amongst these, PTEN, BRCA2, CHEK2, EPCAM, KRAS, and TP53 showed significant discriminative power in distinguishing DM from healthy controls, indicating their potential biomarkers of malignancy risk in DM. These findings shed light on the molecular connection between DM and cancer, suggesting that muscle tissue in patients with DM might be enriched in genes related to various types of cancer. Such a molecular phenotype could provide new insights into devising strategies for malignancy surveillance and early detection in these DM patients, with the aim of improving clinical outcomes. These biomarkers now need to be validated in larger patient cohorts, as well as their role in the guidance of personalized cancer prevention and treatment strategies for patients with dermatomyositis.
REFERENCES: [1] Bachireddy, P., Burman, J., Ling, A., & Turley, S. J. (2018). Dermatomyositis and Malignancy Risk: A Retrospective Cohort Study. Journal of Clinical Oncology , 36(22), 2395-2403.
[2] Fiorentino, D. F., Chung, L. S., & Zwerner, J. P. (2016). The association of cancer with dermatomyositis and polymyositis. Current Rheumatology Reports , 18(2), 1-7.
Acknowledgements: We’d like to thank the Mexican National Council of Humanities, Science and Technology (CONAHCYT) for providing the academic grant that made this research possible. Grant ID: PCC-2022-320697.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (