

Background: Gout is the most common inflammatory arthritis, with a rising incidence and prevalence worldwide. Despite effective treatments, refractory flares are problematic leading to loss of quality of life and work productivity. Current therapies can be limited by comorbidities such as chronic kidney disease and/or adverse effects such as diarrhoea. Therefore, there is a clinical need for additional acute gout treatment options.
Objectives: Interleukin-13 (IL-13) was suggested to play a role in gout pathogenesis. Inhibitors of this cytokine, such as Dupilumab, are licensed for atopic diseases and may have repurposing potential. Natural variation in the gene that encodes a drug target can offer insight into its potential efficacy. Since genetic variants are randomly allocated at conception, genetic instrumental variable analyses, or Mendelian randomization (MR), are typically more robust against biases in traditional pharmacoepidemiologic designs. We used MR to investigate whether genetically proxied IL-13 inhibition influences urate levels or gout risk.
Methods: IL-13 inhibition is recognised to influence eosinophil count in clinical studies; we therefore used this as the downstream biomarker of IL-13 perturbation. We instrumented IL-13 using a missense (protein coding) variant within the IL13 gene that is associated with circulating eosinophil count at genome-wide significance (p<5x10 -8 ) in a study of 563,946 individuals [1]. Genetic association data for gout were derived from 100,671 cases and 2,106,212 controls by the Global Gout Genetics Consortium [2] and replicated first using 2115 cases and 67259 controls from the Global Urate Genetics Consortium [3] and then using 1,699 cases and 216,239 cases in the FinnGen study [4]. Data for serum urate levels were taken from 110,347 individuals [3]. Asthma was used as a positive control (56,167 cases and 352,255 controls) [5]. The Wald ratio or inverse variance weighted methods estimated causal effects, scaled to per standard deviation reduction in eosinophil count. We applied colocalization to assess genetic confounding.
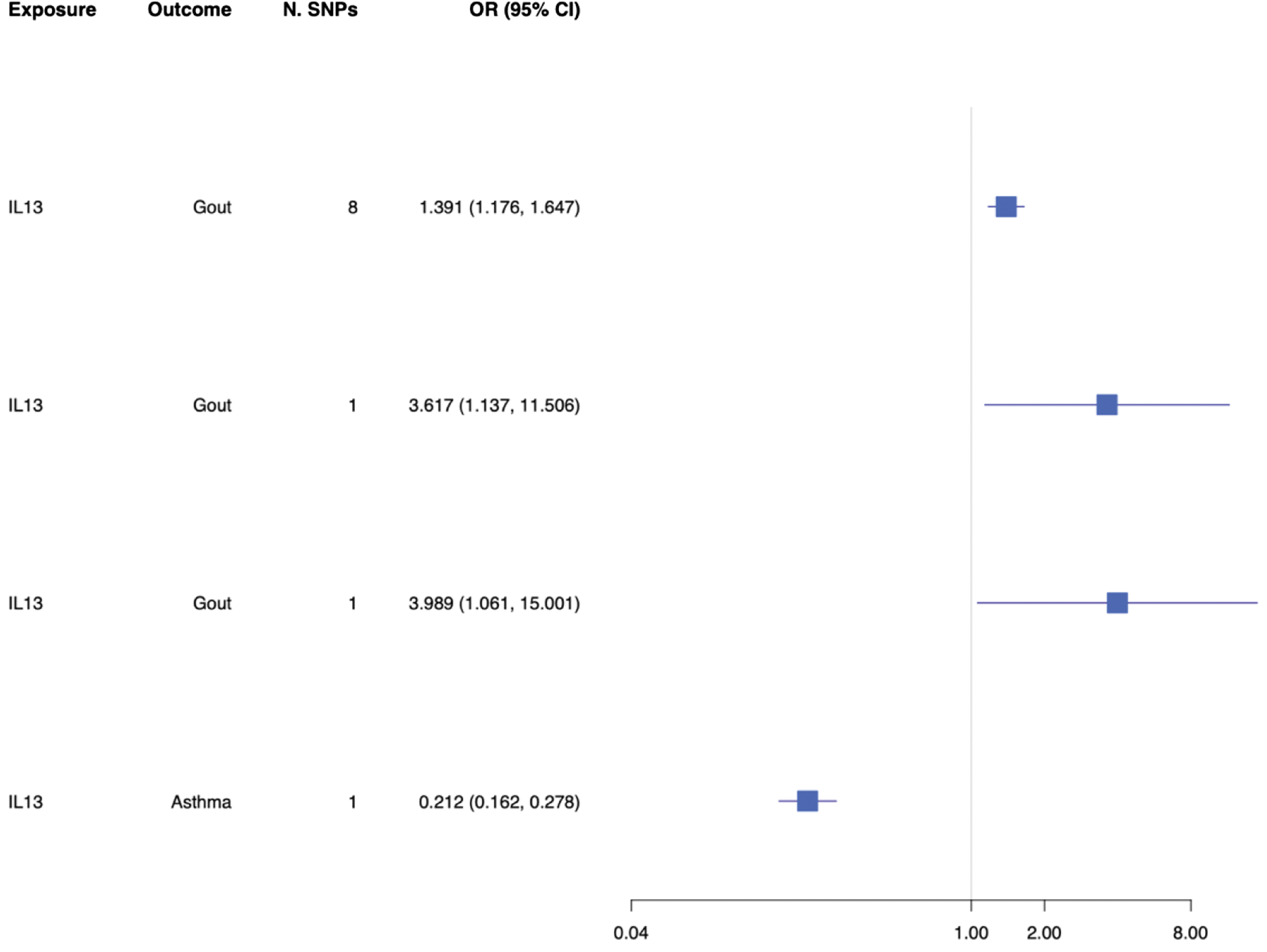
Results: One missense variant, rs20541, was selected to proxy IL-13 inhibition. Genetically proxied IL-13 inhibition was associated with increased gout risk (Odds Ratio 1.65; 95% Confidence Interval [CI] 1.18 to 1.65) which was replicated across independent data sets (Figure 1), but not with urate levels (Beta 0.12, 95% CI -0.09 to 0.33). Colocalisation for the primary analysis showed a conditional probability of 89.9% that results were explained by a shared causal variant.
Conclusion: Our results suggest that the IL-13 pathway is implicated in gout pathophysiology independent of urate levels. However, genetically proxied IL-13 inhibition appears to increase the risk of gout. Further studies are required to characterise gout risk in those with hyperuricaemia and initiating IL-13 inhibitors.
REFERENCES: [1] Vuckovic, D., Bao, E. L., Akbari, P., Lareau, C. A., Mousas, A., Jiang, T., et al. (2020). The polygenic and monogenic basis of blood traits and diseases.
Cell, 182
(5), 1214–1231.e11.
[2] Major, T. J., Takei, R., Matsuo, H., et al. (2024). A genome-wide association analysis reveals new pathogenic pathways in gout.
Nature Genetics, 56
(12), 2392–2406.
[3] Köttgen, A., Albrecht, E., et al. (2013). Genome-wide association analyses identify 18 new loci associated with serum urate concentrations.
Nature Genetics, 45
(2), 145–154.
[4] IEU Open GWAS Project. Dataset:
finn-b-GOUT_STRICT
. Available at:
[5] Valette, K., et al. (2021). Prioritization of candidate causal genes for asthma in susceptibility loci derived from UK Biobank.
Communications Biology, 4
(1), 700.
Forest plot demonstrating the odds ratio with 95% confidence interval for the mendelian randomization assessment of the association between genetically proxied interleukin-13 (IL-13) inhibition with gout and asthma.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (