

Background: Systemic lupus erythematosus (SLE) is a complex inflammatory disease characterized by the development of autoreactive cell and a breakdown of self-tolerance. Natural Killer (NK) cells, pivotal players of the innate immune system, exhibit reduced numbers, diminished cytotoxicity, and impaired cytokine production in SLE patients [1, 2]. Although the molecular mechanisms underlying NK cell dysfunction in SLE remain poorly understood, mitochondrial dynamics appear fundamental in governing the immune cell function [3, 4].
Objectives: To characterize the immunometabolic abnormalities, related to mitochondrial homeostasis, in NK cells from patients with SLE and explore potential strategies for their restoration.
Methods: Mitochondrial function, including membrane potential, mitochondrial mass, superoxide levels, and lysosomal acidification, was assessed using flow cytometry. Transmission electron microscopy (TEM) was employed to examine mitochondrial structure. Flow cytometry functional assays were performed in conjunction with mitochondrial function assays. Quantitative proteomic analysis focused on proteins related to mitochondrial homeostasis and GO (gene ontology) enrichment analysis was performed. qPCR was used to assess the relative abundance of total and cytosolic mitochondrial DNA (mtDNA). RT-qPCR was carried out to examine the expression of mitophagy-related genes.
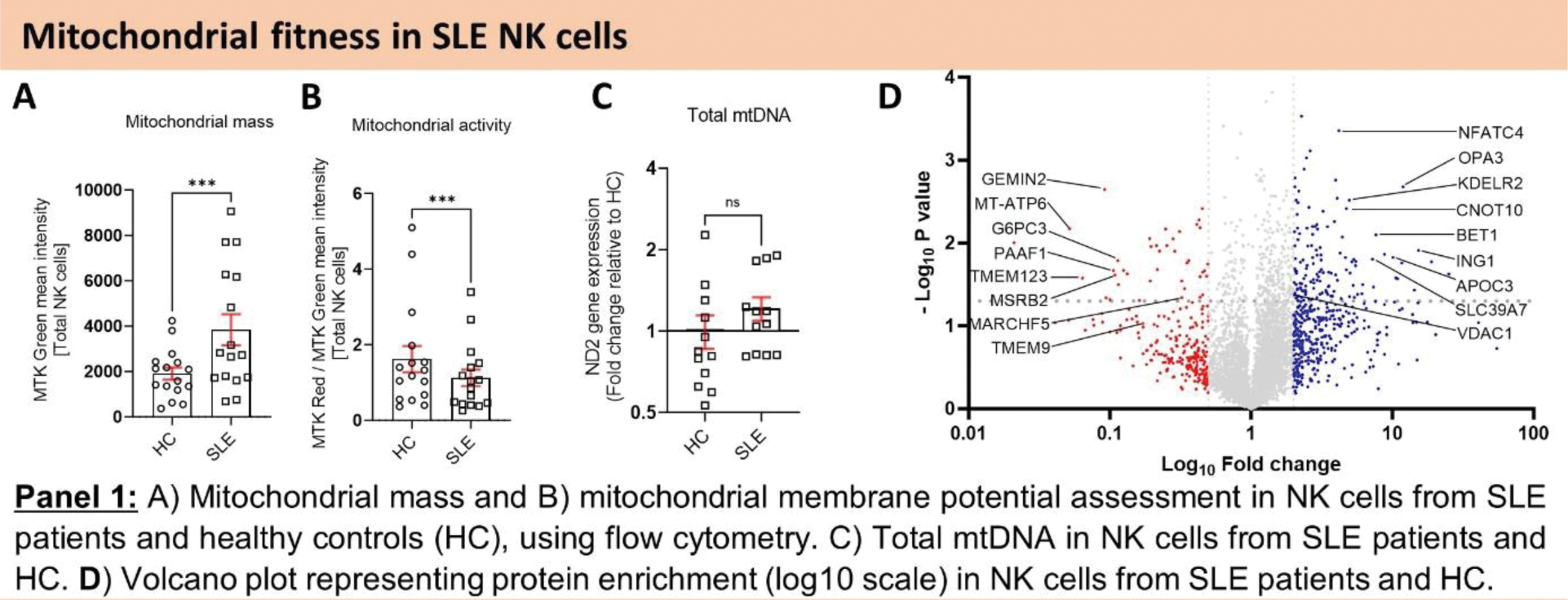
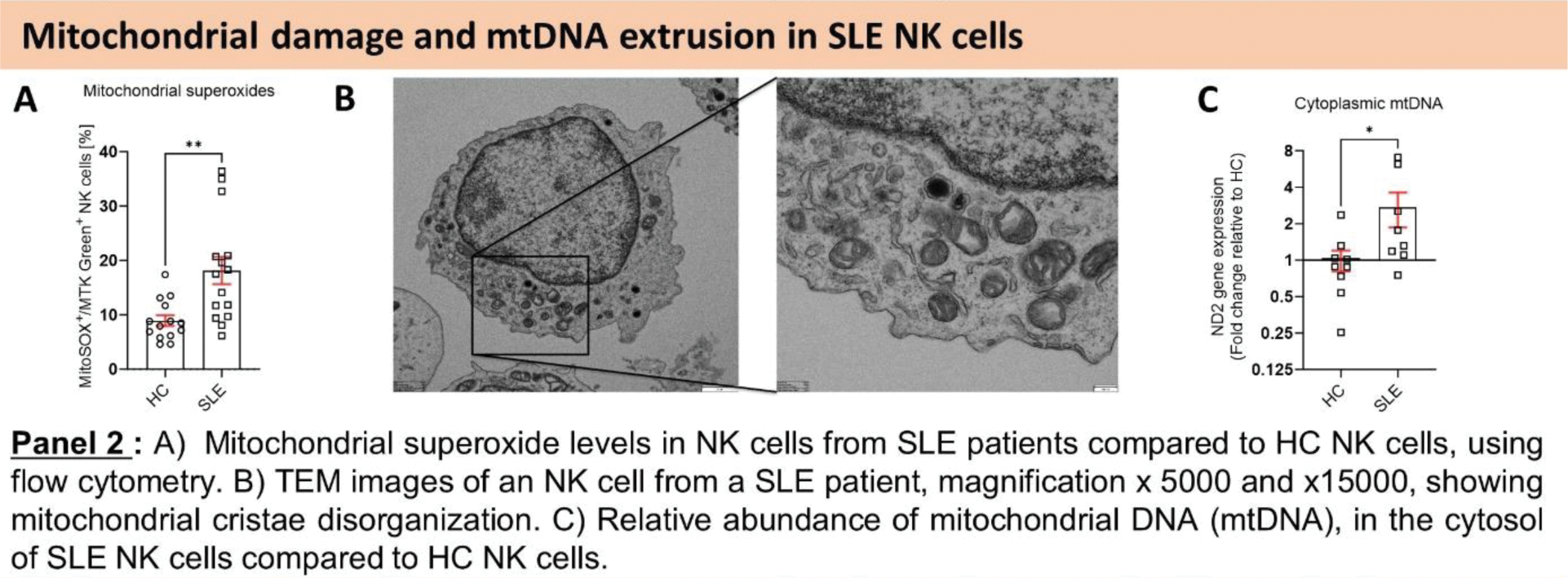
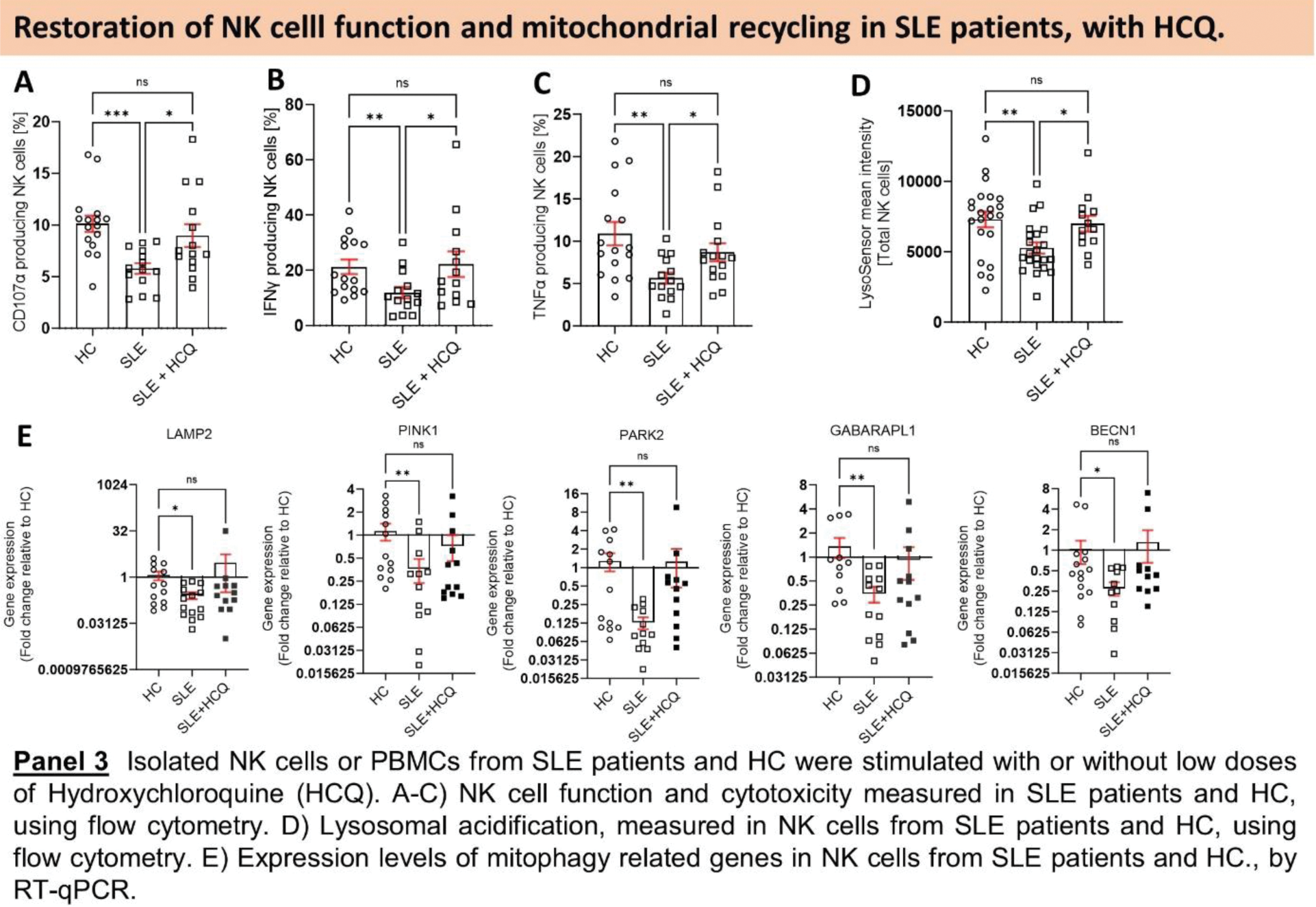
Results: First, we assessed the overall mitochondrial health of SLE NK cells. Our findings indicated an elevation in mitochondrial mass coupled with a reduction in mitochondrial activity (Figure 1.A, B), suggesting an accumulation of dysfunctional mitochondria in SLE NK cells. Quantitative proteomic analysis revealed a reduction in proteins critical for mitochondrial clearance and GO enrichment analysis demonstrated an upregulation of pathways associated with response to DNA damage and DNA metabolic process, in SLE NK cells (Figure 1.D). Furthermore, these SLE NK cell mitochondria exhibited elevated superoxide levels (Figure 2.A). TEM analysis provided visual evidence of cristae disorganization in the mitochondria of SLE NK cells (Figure 2.B). qPCR analysis detected an accumulation of mtDNA in the cytosol of SLE NK cells (Figure 2.C). Our flow cytometry analysis demonstrated that while the number of lysosomes in SLE NK cells was normal, their pH was more alkaline compared to healthy controls (Figure 3.D). To elucidate the molecular mechanism driving the increased mitochondrial mass in SLE NK cells, we examined the effect of V-ATPase inhibition using Bafilomycin A1. Bafilomycin A1 increased mitochondrial mass by inhibiting lysosomal trafficking and heightened mtDNA extrusion, in healthy NK cells. These results suggest a link between the presence of enlarged dysfunctional mitochondria and their recycling. To decipher this dysfunction in mitochondrial recycling in SLE NK cells, we evaluated the effect of Hydroxychloroquine (HCQ) on the lysosomal and cell function in parallel to mitophagy genes expression. Our in vitro exhibited HCQ ‘s ability to restore cell function (Figure 3.A-C), lysosomal acidification (Figure 3.D) and mitophagy genes expression including LAMP2, PINK1, PARK2, PIK3C3, GAPARAPL1, BECN1 (Figure 3.E), in NK cells from SLE patients.
Conclusion: Our study has revealed a wide spectrum of alterations affecting the mitochondrial homeostasis in SLE NK cells, characterized by an accumulation of enlarged hyperpolarized mitochondria, with disrupted cristae. Mitochondria in SLE NK cells exhibited heightened superoxide levels, mtDNA extrusion and downregulated expression of mitophagy-related genes. Our findings strongly indicate that impaired mitochondrial turnover majorly contributes to the dysfunction of SLE NK cells. Addressing these abnormalities, specifically targeting the restoration of mitophagy-related genes expression levels, the lysosomal and cell function, with small doses of HCQ, holds promise as a therapeutic strategy. HCQ emerges as potential candidates to mitigate these perturbations in SLE NK cells and sheds light on a previously undisclosed mechanism underlying the beneficial effects of HCQ in SLE patients.
REFERENCES: [1] Spada R, Rojas JM, Barber DF. Recent findings on the role of natural killer cells in the pathogenesis of systemic lupus erythematosus. J Leukoc Biol. 2015;98(4):479-87.
[2] Park Y-W, Kee S-J, Cho Y-N, Lee E-H, Lee H-Y, Kim E-M, et al. Impaired differentiation and cytotoxicity of natural killer cells in systemic lupus erythematosus. Arthritis & Rheumatism. 2009;60(6):1753-63.
[3] Caza TN, Fernandez DR, Talaber G, Oaks Z, Haas M, Madaio MP, et al. HRES-1/Rab4-mediated depletion of Drp1 impairs mitochondrial homeostasis and represents a target for treatment in SLE. Ann Rheum Dis. 2014;73(10):1888-97.
[4] Lu Z, Tian Y, Bai Z, Liu J, Zhang Y, Qi J, et al. Increased oxidative stress contributes to impaired peripheral CD56(dim)CD57(+) NK cells from patients with systemic lupus erythematosus. Arthritis Res Ther. 2022;24(1):48.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (