

Background: Psoriatic Arthritis (PsA) is a chronic and complex inflammatory disease with heterogeneous manifestations. Despite the advances in treatment that have led to improved management of PsA, disease modifying drug (DMARD) response rates remain relatively low [1]. A promising approach to improve PsA treatment responses is incorporating predictive biomarkers to better tailor treatment to individual patients (personalised treatment) [2].
Objectives: We aim to find transcriptomic and proteomic markers that can predict a patient’s treatment outcome and, consequently, to develop a prediction model to aid in treatment choice for PsA patients.
Methods: Baseline data from the TOFA-PREDICT trial discovery cohort including DMARD-naïve PsA patients and PsA patients that previously failed DMARD therapy (DMARD-failure) [3] were used. DMARD-naïve patients were randomised to either tofacitinib (n=20) or methotrexate (n=20), and DMARD-failure patients were randomised to receive tofacitinib (n=20) or etanercept (n=20). Treatment response was defined as reaching Minimal Disease Activity (MDA) after 16 weeks of treatment. RNA sequencing in CD4 T cells was performed using the Illumina NovaSeq6000 sequencing platform. Olink Proteomics technology was used for proteomic profiling of 368 proteins in serum samples (cardiovascular, cardiometabolic, inflammation, and metabolism panels). Transcriptomic and proteomic predictive markers were selected using XGBoost and/or sPLS-DA in the total cohort and in the treatment subgroups. Subsequently, we developed a transcriptomic and a proteomic prediction model for response to treatment using Ridge regression based on the selected transcriptomic or proteomic markers, previously selected baseline clinical variables, and treatment group (tofacitinib, methotrexate or etanercept). Interaction terms between the treatment group predictors are included in the models to allow treatment-specific prediction. Prediction models were optimised and evaluated using double-loop leave-one-out cross-validation.
Results: The Health Assessment Questionnaire (HAQ), Body Mass Index (BMI), DMARD history, and Disease Activity in PSoriatic Arthritis (DAPSA) score were used as clinical covariates for the prediction models. 7 gene transcripts and 12 proteins were selected as predictive markers for, respectively, the transcriptomics-prediction model and the proteomics-prediction model. Final models showed an AUC-ROC 0.89 and 0.78 (Figure 1a and 1b). To demonstrate the potential for treatment-specific prediction the probability of achieving MDA was calculated for tofacitinib and standard treatment (ethanercept or methotrexate) using both prediction models. The predicted individualised treatment effects comparing tofacitinib to standard treatment only varied minimally between patients (less than 5% for both models).
(A ) ROC curve of the transcriptomics-prediction model. The λ was evaluated using nested leave-one-out cross-validation. The 7 gene-transcript included in the model are ENSG00000196557, ENSG00000232472, ENSG00000244398, ENSG00000215105, ENSG00000170584, ENSG00000167720, and ENSG00000232587. (B ) ROC curve of the proteomics-prediction model. The λ was evaluated using nested leave-one-out cross-validation. The 12 proteins included in the model are: FCGR2A, QPCT, VCAM1, OPN, CA13, CD5, FGF-19, ANGPT2, ALDH1A1, GHRL, QPCT, and PSP-D.
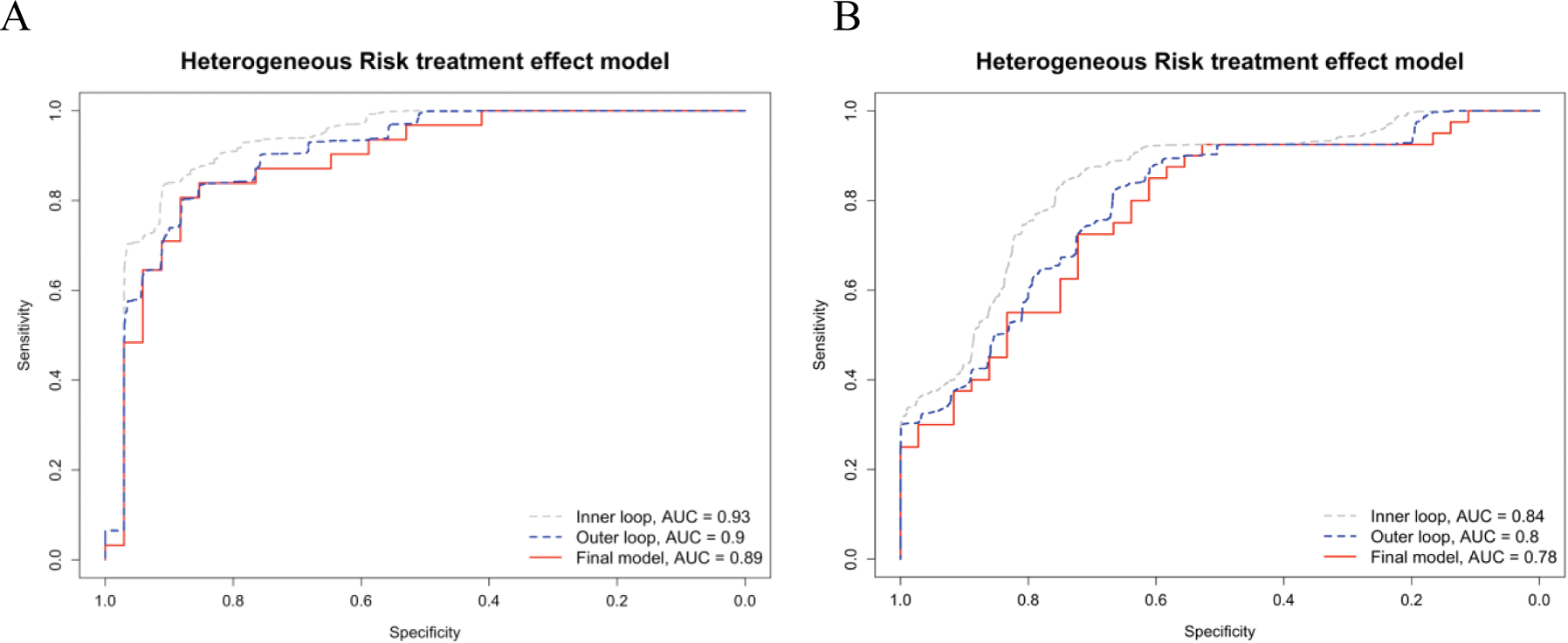
Conclusion: The combination of clinical information with transcriptomic or proteomic markers can be used for the prediction of response to treatment relatively well, especially for the transcriptomics-based prediction model. Unfortunately, the variation in predicted individual treatment effects was small and is unlikely to aid in individual treatment decisions. Therefore, we are currently evaluating a different heterogeneous treatment effect model, as well as an integrated multi-omics approach for predicting individualized treatment effects toward personalised treatment in PsA.
REFERENCES: [1] Zardin-Moraes M, Silva ALFA da, Saldanha C, et al. Prevalence of Psoriatic Arthritis Patients Achieving Minimal Disease Activity in Real-world Studies and Randomized Clinical Trials: Systematic Review with Metaanalysis. J Rheumatol. 2020;47(6):839-846. doi:10.3899/jrheum.190677.
[2] Gurke R, Bendes A, Bowes J, et al. Omics and Multi-Omics Analysis for the Early Identification and Improved Outcome of Patients with Psoriatic Arthritis. Biomedicines. 2022;10(10):2387. doi:10.3390/biomedicines10102387.
[3] Kleinrensink, Nienke J., et al. “TOFA-PREDICT study protocol: a stratification trial to determine key immunological factors predicting tofacitinib efficacy and drug-free remission in psoriatic arthritis (PsA).” BMJ open 12.10 (2022): e064338.
Acknowledgements: This work was supported by Pfizer (New York, New York, USA). The collaboration project is co-funded by the PPP Allowance made available by Health∼Holland, Top Sector Life Sciences & Health, to stimulate public–private partnerships (grant number: LSHM17074).
Disclosure of Interests: Mieke L. M. Bentvelzen: None declared, Said el Bouhaddani: None declared, Julia Spierings: None declared, Harald E. Vonkeman paid consultant for (pharmaceutical) companies Abbvie, Novartis, Pfizer, UCB, Johnson and Johnson, financial grants from (pharmaceutical) companies: Galapagos and Boehringer Ingelheim, Shasti C. Mooij: None declared, Lydia G. Schipper: None declared, Amin Herman: None declared, Simon C. Mastbergen: None declared, Paco M.J. Welsing: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (