

Background: In patients with a usual interstitial pneumonia pattern, differentiating Idiopathic Pulmonary Fibrosis (IPF) from Rheumatoid Arthritis-associated Interstitial Lung Disease (RA-ILD) is a clinically significant but challenging task. While both conditions share overlapping imaging features, their management and prognostic implications differ substantially. Traditional methods, relying on visual assessment of high-resolution computed tomography (HRCT) scans, are dependent on the operator’s expertise and can lead to diagnostic uncertainty, particularly in cases with similar radiologic presentations. Advances in radiomics and machine learning have opened new possibilities for overcoming these limitations by extracting quantitative features from imaging data. Radiomics, by analyzing shape, texture, and intensity patterns, provides an objective and high-dimensional characterization of lung abnormalities. When coupled with machine learning algorithms, radiomics has the potential to distinguish between subtle disease phenotypes with high accuracy [1].
Objectives: This study aimed to evaluate the usefulness of radiomic features extracted from HRCT scans in differentiating IPF from RA-ILD in patients with a UIP pattern and to assess model performance while controlling for disease severity.
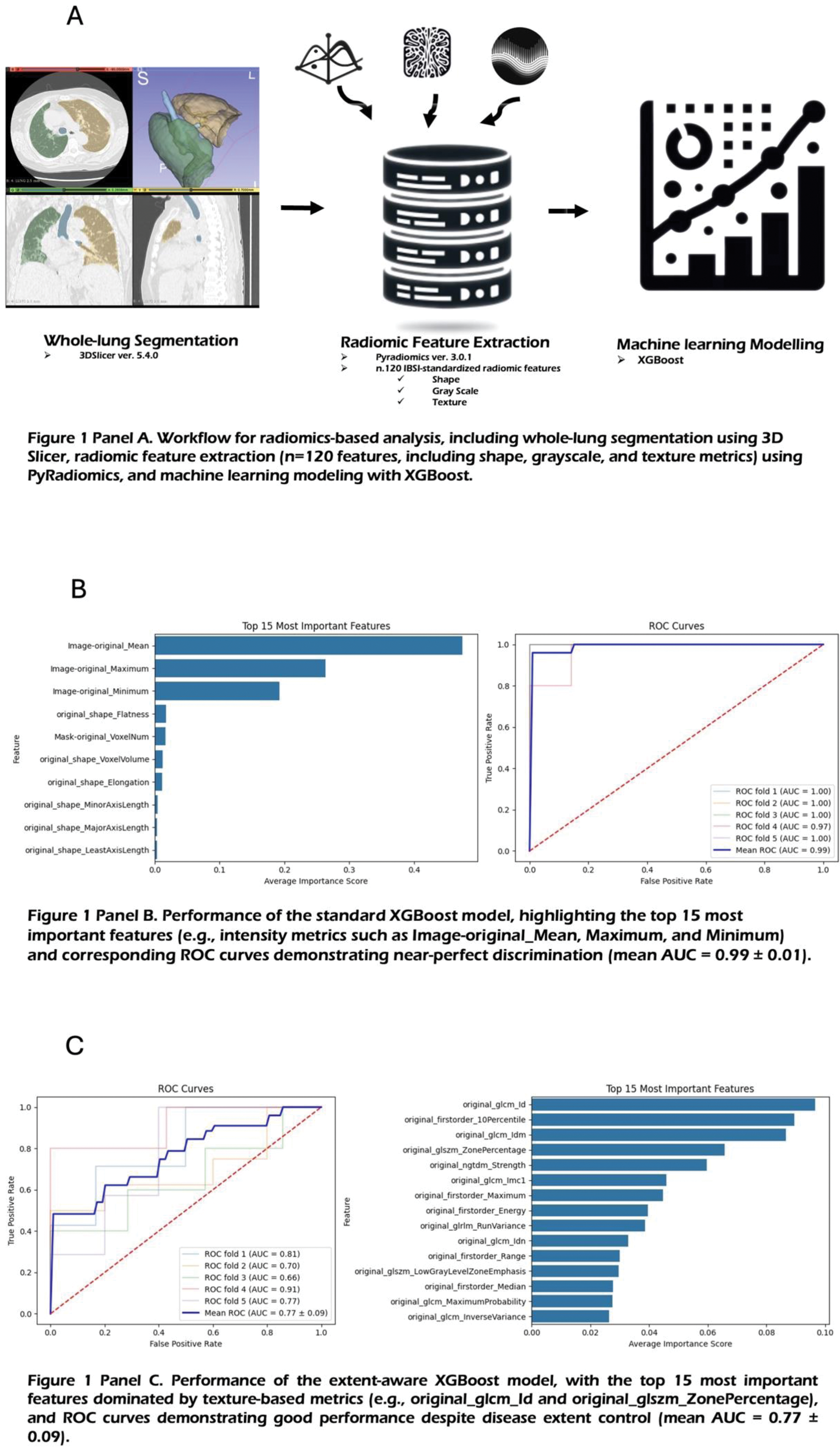
Methods: This retrospective study analyzed HRCT scans from 72 patients (42 IPF, 30 RA-ILD) confirmed to have a UIP pattern. Lung volumes were segmented using 3DSlicer (version 5.4.0), and 120 radiomic features were extracted using PyRadiomics (version 3.0.1, Figure 1 Panel A, see
Standard Model: All features were included to assess overall discriminative performance.
Extent-Aware Model: To control for disease severity, radiomic features related to volume were excluded, and a disease extent proxy (mesh volume) was incorporated into the analysis.
Performance was assessed using 5-fold cross-validation, with model accuracy quantified by the area under the receiver operating characteristic curve (AUC). Statistical analyses were complemented by survival analysis to examine the prognostic implications of accurate classification.
Results: The study cohort consisted of 42 IPF patients and 30 RA-ILD patients. The IPF group had a male predominance (73.81%, 31/42) and a median age of 78 (IQR 8) years, whereas the RA-ILD group had a female predominance (63.33%, 19/30) and a median age of 72 (IQR 12.5). The Standard XGBoost Model achieved near-perfect discrimination (AUC 0.99 ± 0.01, Figure 1 Panel B). First-order intensity features were the most important contributors to the model’s performance, including:
Image-original_Mean: Average intensity of the segmented lung volume.
Image-original_Maximum: Highest intensity voxel value.
Image-original_Minimum: Lowest intensity voxel value.
In the Extent-Aware Model, performance remained robust (AUC 0.77 ± 0.09, Figure 1 Panel C), with texture-based features emerging as key discriminators. These included:
original_glcm_Id: Inverse difference, reflecting texture homogeneity.
original_firstorder_10Percentile: Intensity value below which 10% of voxel values fall.
original_glszm_ZonePercentage: Proportion of homogeneous zones relative to the total lung volume.
Survival analysis revealed significantly worse outcomes in the IPF cohort (median survival: 5.5 months, 14 events) compared to the RA-ILD cohort (median survival: 29.5 months, 13 events). These findings underscore the clinical importance of accurate differentiation, as therapeutic strategies and prognoses differ substantially.
Conclusion: Radiomics analysis demonstrates strong potential in differentiating IPF from RA-ILD with a UIP pattern. The near-perfect discrimination achieved by intensity-based features in the standard model highlights the utility of radiomics in clinical practice. Importantly, the extent-aware model’s reliance on texture-based features provides evidence that radiomics can distinguish these conditions independently of disease severity, offering a robust approach for early diagnosis. Given the more than five-fold difference in median survival, accurate differentiation has significant implications for treatment planning and management. Future studies should explore external validation in larger, multicenter cohorts and assess the integration of radiomics into routine clinical workflows to optimize patient outcomes.
REFERENCES: [1] Venerito V et al. (2023) doi: 10.3389/fmed.2022.1069486.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (