

Background: Artificial Intelligence (AI) has shown great potential to improve the identification of drug interactions, especially in patients with multiple treatments, such as rheumatology patients. Tools like ChatGPT-3.5, ChatGPT-4, Gemini, and Copilot can offer quick assistance, but their accuracy compared to clinically validated databases like Lexicomp has not been exhaustively evaluated in a real clinical context. This study compares these AI platforms in detecting drug interactions in rheumatology.
Objectives: To evaluate the accuracy, sensitivity, and specificity of ChatGPT-3.5, ChatGPT-4, Gemini, Gemini Adv, and Copilot compared to the Drugs.com database and using Lexicomp as the gold standard, and to analyze the impact of using specific prompts on their performance.
Methods: The study included 520 drug interaction scenarios, generated from the 10 most prescribed drugs in rheumatology and the 50 most common drugs in the Community of Madrid in 2023. Interactions were classified by clinical relevance (grades 0 to 4). The results from each tool were compared with Lexicomp, the reference standard, and metrics such as accuracy, sensitivity, specificity, and area under the curve (AUC) were calculated.
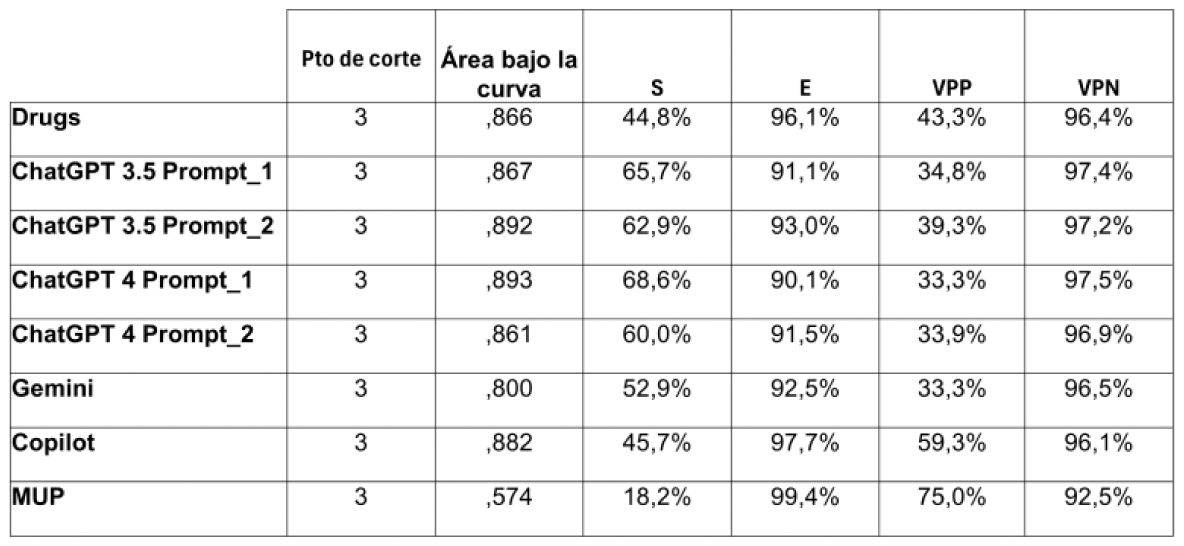
Results: Of the 520 interactions analyzed, 35 were classified as clinically relevant by the Lexicomp database (grades 3 and 4). All AI tools showed AUC values above 0.8, with ChatGPT-4 being the best performer with an AUC of 0.882. A similar result was obtained with the Drugs.com database (AUC = 0.864). The only exception was the MUP tool, which achieved an AUC of 0.574 (see Figure 1, ROC Curves). In terms of sensitivity, ChatGPT-4 achieved 81%, followed by Gemini (78%), ChatGPT-3.5 (75%), and Copilot (73%), while MUP reached only 58%. Specificity was high across all platforms, with ChatGPT-4 achieving 88%, followed by Gemini (85%), ChatGPT-3.5 (83%), and Copilot (81%). MUP had a specificity of 71% (see Table 1 for detailed sensitivity, specificity, and other parameters).
Conclusion: AI tools, especially ChatGPT-4, showed performance comparable to the reference standard in identifying clinically relevant drug interactions, with high AUC, sensitivity, and specificity values. The use of specific prompts significantly improved their accuracy. The MUP tool, despite its advances, showed inferior performance and requires optimization for clinical application.
REFERENCES: NIL.
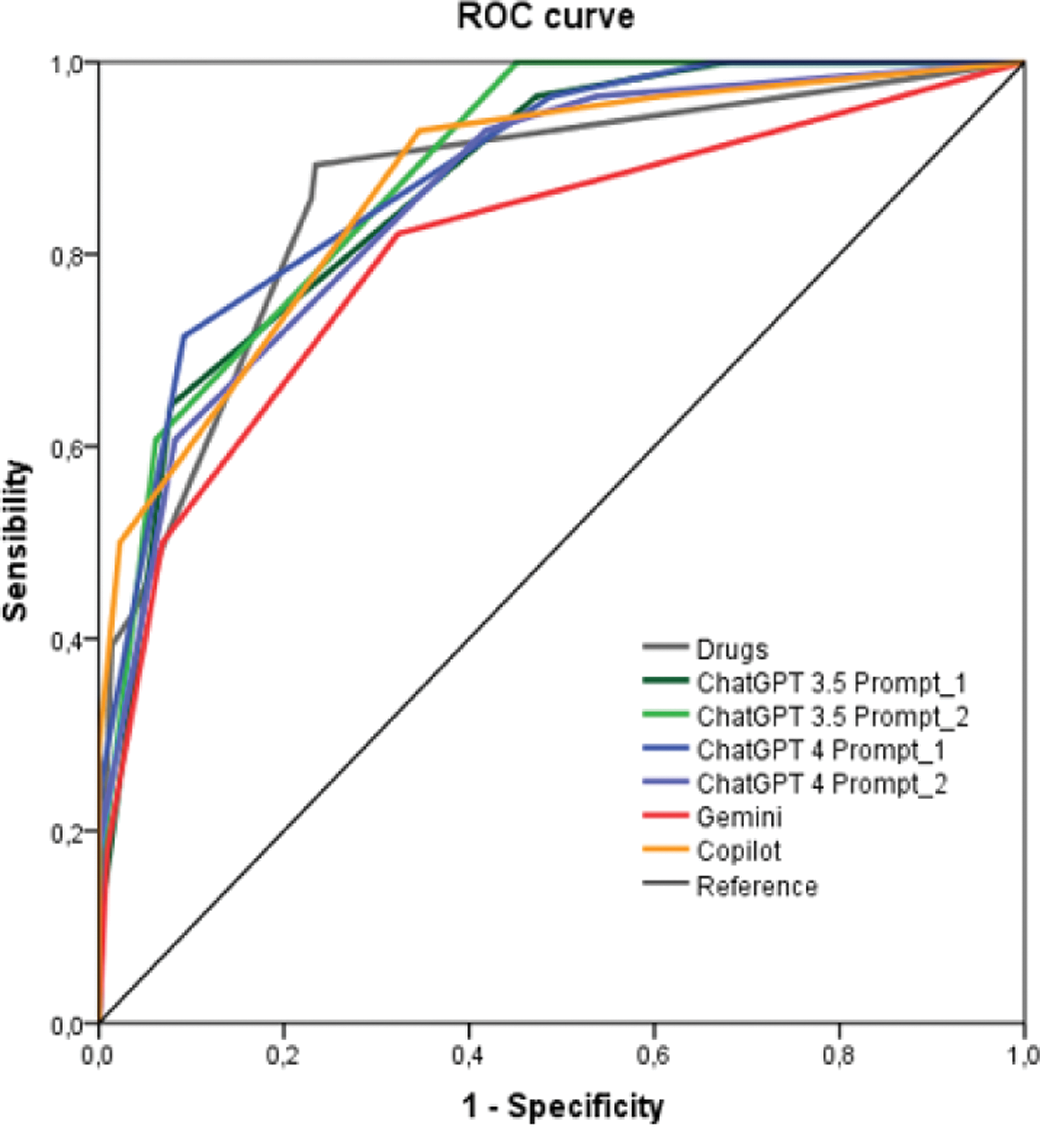
Table 1.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (