

Background: Systemic Lupus Erythematosus (SLE) is a complex autoimmune disease characterized by heterogeneous clinical manifestations and a pathogenesis influenced by genetic, hormonal, and environmental factors. Antinuclear antibodies (ANAs) play a pivotal role in the diagnosis and monitoring of SLE, although their precise clinical relevance remains incompletely understood. In this study, machine learning (ML) techniques were utilized to identify clinical subgroups among patients positive for anti-nucleosome antibodies, offering a novel approach for stratifying patients and enabling personalized management of SLE.
Objectives: 1. To identify clinical subgroups among SLE patients with positive anti-nucleosome antibodies based on clinical and laboratory data. 2. To stratify patients according to disease prognosis severity, with the aim of optimizing clinical evaluation and follow-up.
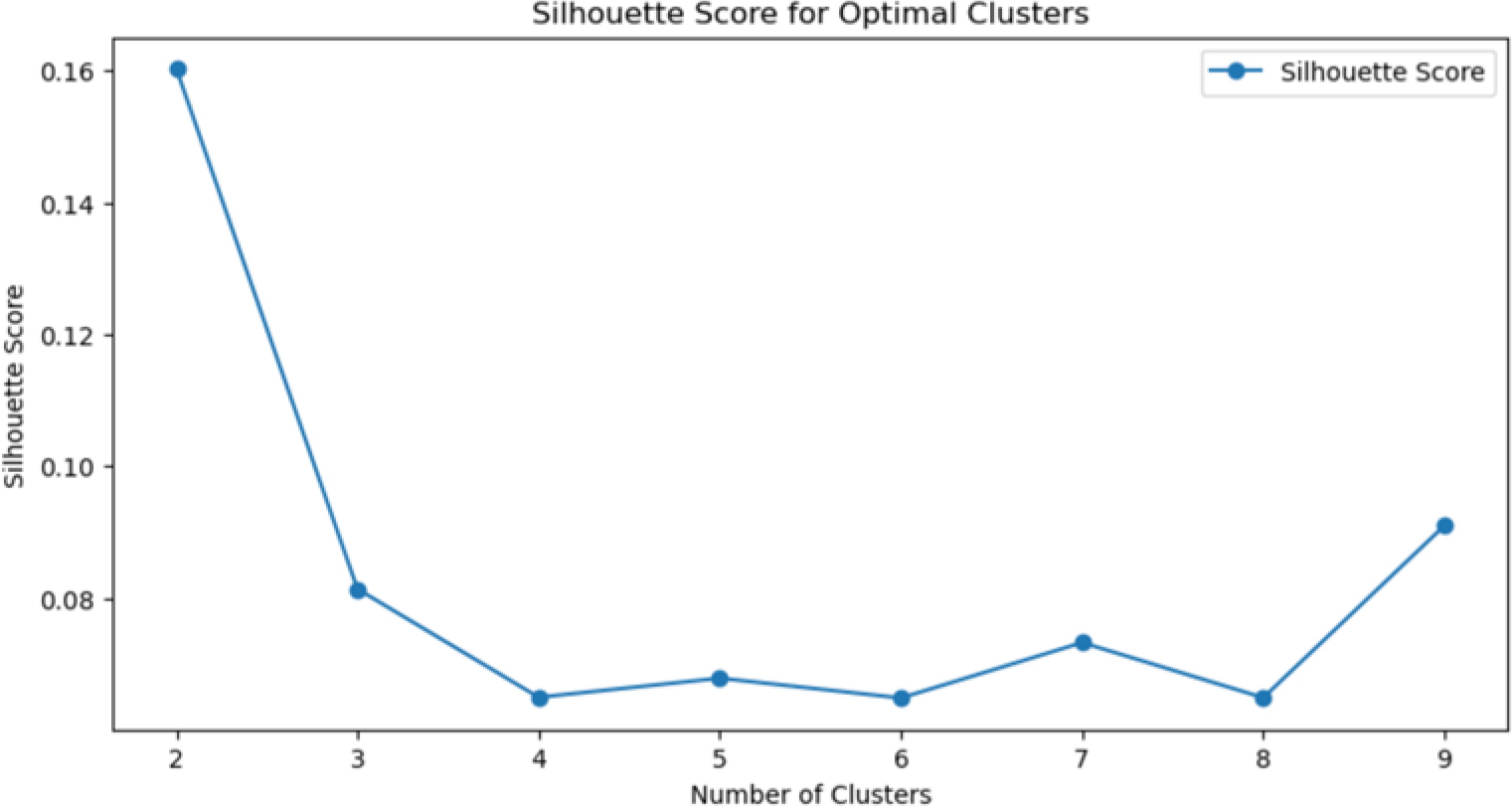
Methods: Retrospective data were collected from a tertiary hospital in Barcelona (2009–2024), including patients with specific SLE-associated antibodies, resulting in a final cohort of 111 individuals. Epidemiological, clinical, and laboratory parameters were analyzed, with a specific focus on anti-nucleosome antibodies. Data preprocessing involved z-score normalization and median imputation for missing data. The K-Means clustering algorithm was selected, and the optimal number of clusters was determined using the elbow method and silhouette analysis, with Python-based tools. Results were visualized using t-SNE plots.
Results: The cohort (84.68% female; mean age 32.5 years) exhibited a predominance of joint-related symptoms, with arthralgia (54.05%) and arthritis (35.14%) being the most common. Among 39 patients positive for anti-nucleosome antibodies, clustering analysis identified two distinct subgroups (Figure 1):
Cluster 0 (C0): Patients with milder symptoms, reduced systemic involvement, and lower immune activity, with a notable prevalence of Raynaud’s phenomenon.
Cluster 1 (C1): Patients with higher disease activity, systemic involvement, and elevated immune markers, including anti-Ro52 and anti-C1q antibodies.
An unexpected relation was identified between Raynaud’s phenomenon and reduced systemic disease progression in patients positive for anti-nucleosome antibodies, suggesting a potential protective role that warrants further investigation (Table 1).
Conclusion: Clustering techniques have demonstrated their potential to identify distinct clinical phenotypes in patients with heterogeneous diseases such as SLE. In this study, Raynaud’s phenomenon was associated with milder disease activity. This finding underscores the need for future studies with multicenter cohorts and larger sample sizes to validate these observations. Relevance and Research Perspectives : The significance of this work lies in sharing the methodology and highlighting the potential of innovative tools to accurately classify patients and optimize follow-up strategies. Moreover, this study aims to inspire both young and experienced rheumatologists to embrace this scientific paradigm shift and collaborate to advance the field of rheumatology.
This graph displays the silhouette coefficient values calculated through cluster analysis, using the elbow method as a reference. The silhouette coefficient is a quantitative metric that evaluates the internal cohesion of data within clusters and their separation from other clusters. According to the results presented, the maximum silhouette coefficient value is achieved with two clusters, indicating that this configuration represents the optimal number of clusters for the dataset analyzed.
Mean values of clinical characteristics according to the cluster analysis for the identified groups: C0 and C1. Negative values reflect a lower association or prevalence, while positive values indicate a higher association. Although the differences between clusters are generally subtle, Raynaud’s phenomenon stands out in C0 (mean: 1.10) as a potential distinctive marker.
| Feature | Mean values (C0) | Mean values (C1) |
|---|---|---|
| Fever | -0.34 | 0.10 |
| Lymphadenopathy | -0.34 | 0.10 |
| Sicca syndrome | -0.43 | 0.13 |
| Arthralgia | -1.7 | 0.51 |
| Arthritis | -0.93 | 0.28 |
| Raynaud’s syndrome | 1.10 | -0.33 |
| Cutaneous lupus | -0.33 | 0.1 |
| Alopecia | -0.51 | 0.15 |
| Aphthosis | -0.02 | 0.01 |
| Glomerulonephritis I/II | -0.38 | 0.12 |
| Glomerulonephritis III/IV | 0.69 | -0.21 |
| Pleuritis | -0.16 | 0.05 |
| Pericarditis | -0.16 | 0.05 |
| Thrombopenia | 0.76 | -0.23 |
| Neutropenia | -0.23 | 0.07 |
| Lymphopenia | -0.05 | 0.02 |
| Leucopenia | -0.18 | 0.05 |
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: Asier García-Alija Amgen, Berta Paula Magallares GSK, AstraZeneca, Andrea Hernández: None declared, Guillem Verdaguer: None declared, Anna Calvet: None declared, Ana Laiz Abbvie, Johnson & Johnson, UCB, Novartis, UCB, Patricia Moya GSK, Albert Casals Urquiza: None declared, Cesar Díaz-Torné Abbvie, Lilly, Alfasigma, Novartis, UCB, Luís Sainz Comas Johnson & Johnson, Abbvie, Lilly, Alfasigma, Novartis, UCB, Ivan Castellví Boehringer-Ingelheim, GSK, Novartis, Boehringer-Ingelheim, GSK, Johnson & Johnson, Novartis, Boehringer-Ingelheim, Sanofi, GSK, Susana P. Fernandez-Sanchez AstraZeneca y GSK, Julia Bernardez Otsuka, Helena Codes: None declared, Jose Luis Tandaipan Johnson & Johnson, Concepción Pitarch: None declared, Carla Marco Pascual: None declared, Andrea Garcia Guillen: None declared, Maria Àngels Melchor: None declared, Margarita Sihuro: None declared, Sandra Ros: None declared, Lorena Úbeda: None declared, Núria Fernández-Verdés: None declared, Hèctor Corominas: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (