

Background: Rheumatoid arthritis is a chronic inflammatory autoimmune disease characterized by persistent synovitis, leading to joint destruction, functional disability, and systemic complications [1]. The exact pathogenesis is still unclear. Previous studies have found that mitochondria play a key role in bone metabolic diseases and are key players in the pathogenesis and treatment of RA [2-4]. However, specific mitochondrial-related genes and their role in RA have remained elusive [2, 5]. Therefore, it is necessary to further explore the causal relationship between mitochondria and RA pathogenesis.
Objectives: We employed two-sample Mendelian randomization and colocalization analyses to investigate the associations between mitochondrial-related genes and RA by integrating multi-omics data.
Methods: Summary-level data on mitochondrial gene methylation, expression, and protein abundance were obtained from their respective quantitative trait loci studies. We obtained genetic associations with RA from the GWAS catalog and the FinnGen study. We performed two-sample Mendelian randomization analysis to assess the associations of mitochondrial gene-related molecular features with RA. The Steiger filter test was used to test the causal direction. Colocalization analysis was further conducted to assess whether the identified signal pairs shared a causal genetic variant (Figure 1).
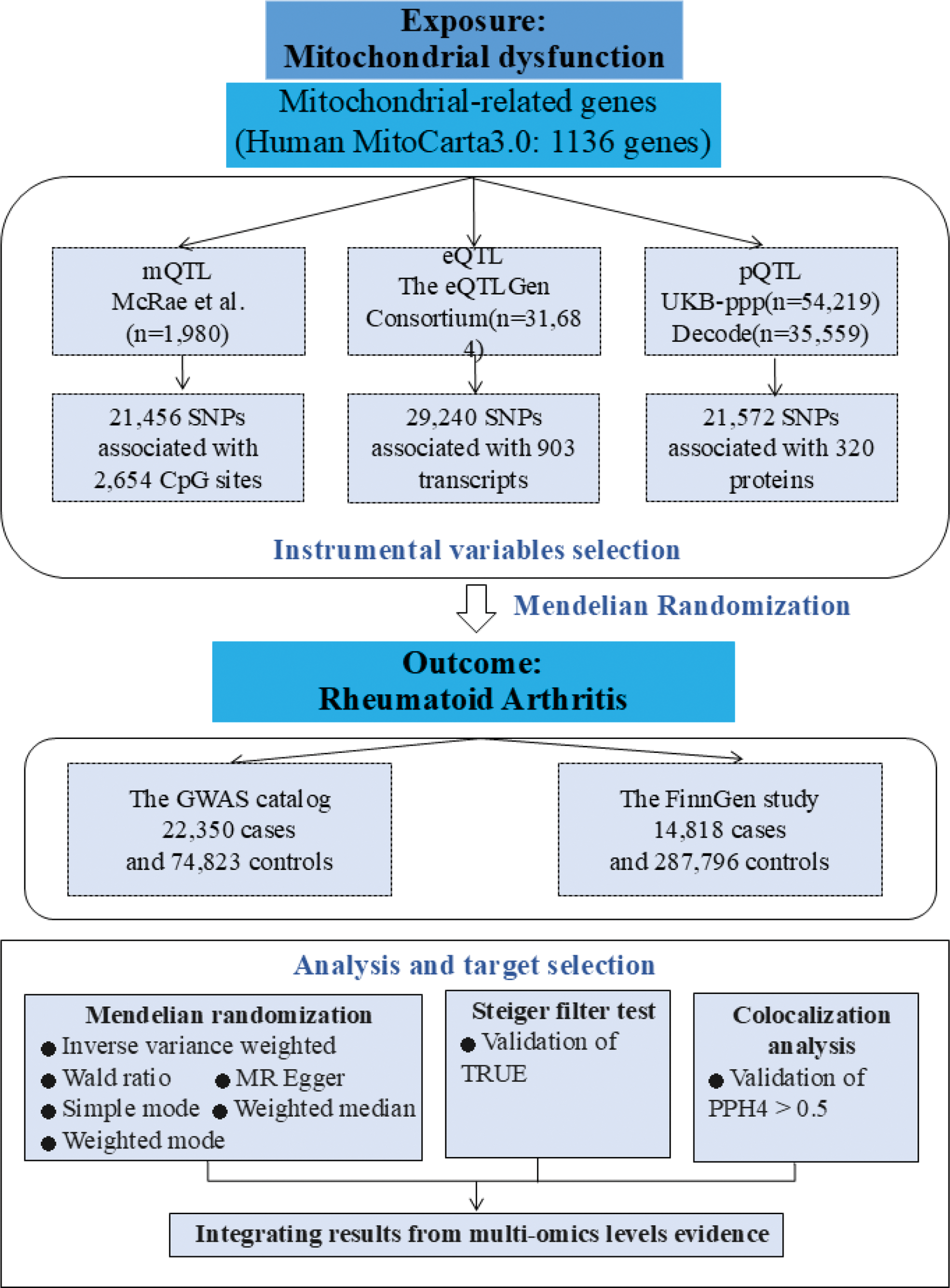
Results: After integrating evidence from multi-omics, we identified the gene CASP3, whose methylation levels (cg12167823, cg21490662), gene expression, and protein levels were inversely associated with the risk of RA. Similarly, a negative association was observed between methylation levels and the risk of RA for TUFM, while a positive association between gene expression and the risk of RA was also identified. Moreover, colocalization analysis found strong evidence (PPH4>0.70) for TUFM at both methylation and gene levels. The methylation level of HSD17B8 (cg02647520) was negatively correlated with the risk of RA; the methylation level of VARS2 (cg02306936) was positively correlated with the risk of RA; the gene expressions of BCL2L11 and SUOX were positively correlated with the risk of RA. Colocalization analysis found strong evidence for the CpG site cg02647520 of gene HSD17B8, the CpG site cg02306936 of gene VARS2, BCL2L11, and SUOX (PPH4>0.70) (Table 1).
The results of two-sample Mendelian randomization analysis and colocalization analysis
| id.outcome | Gene | mQTL | eQTL | pQTL | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ProbeID | p | OR(95%CI) | PPH4 | p | OR(95%CI) | PPH4 | Cohort | p | OR(95%CI) | PPH4 | ||
| finngen_R11_M13_RHEUMA
| CASP3
| cg12167823
| 0.008
| 0.974(0.955-0.993)
|
| 0.049
| 0.963(0.927-1.000)
|
| decode
| 0.013
| 0.909(0.842-0.980)
|
|
Conclusion: This study established a causal relationship between mitochondrial biological function and RA through a multi-omics approach, providing new insights for RA diagnosis and treatment.
REFERENCES: [1] Di Matteo A, Bathon JM, Emery P. Rheumatoid arthritis. Lancet. 2023 Nov 25;402(10416):2019-2033.
[2] Clayton SA, MacDonald L, Kurowska-Stolarska M, Clark AR. Mitochondria as Key Players in the Pathogenesis and Treatment of Rheumatoid Arthritis. Front Immunol. 2021 Apr 29;12:673916.
[3] Becker YLC, Duvvuri B, Fortin PR, Lood C, Boilard E. The role of mitochondria in rheumatic diseases. Nat Rev Rheumatol. 2022 Nov;18(11):621-640.
[4] Wang S, Deng Z, Ma Y, Jin J, Qi F, Li S, Liu C, Lyu FJ, Zheng Q. The Role of Autophagy and Mitophagy in Bone Metabolic Disorders. Int J Biol Sci. 2020 Jul 30;16(14):2675-2691.
[5] Ma C, Wang J, Hong F, Yang S. Mitochondrial Dysfunction in Rheumatoid Arthritis. Biomolecules. 2022 Sep 1;12(9):1216.
Acknowledgements: This project was supported by grants from the Natural Science Foundation of Shanxi Province (No. 20210302123274).
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (