

Background: Interactions among autoimmunity, vasculopathy, and fibrotic tissue remodeling shape the microenvironment and affect the disease outcome in Systemic Sclerosis (SSc). Despite recent advances in scar-forming cell heterogeneity through single-cell RNA sequencing (scRNA-seq), the regulatory network within the fibrogenesis microenvironment remains elusive.
Objectives: We aimed to explore the distinct cell types and their neighbourhood interaction in SSc skin, providing spatiotemporal insights.
Methods: We generated spatially resolved transcriptome maps for healthy and SSc skin based on 10× Visium and Stereo-seq platforms, which was validated using spatial proteomic techniques and time-course scRNA-seq data of SSc mouse and wound-healing reindeer models.
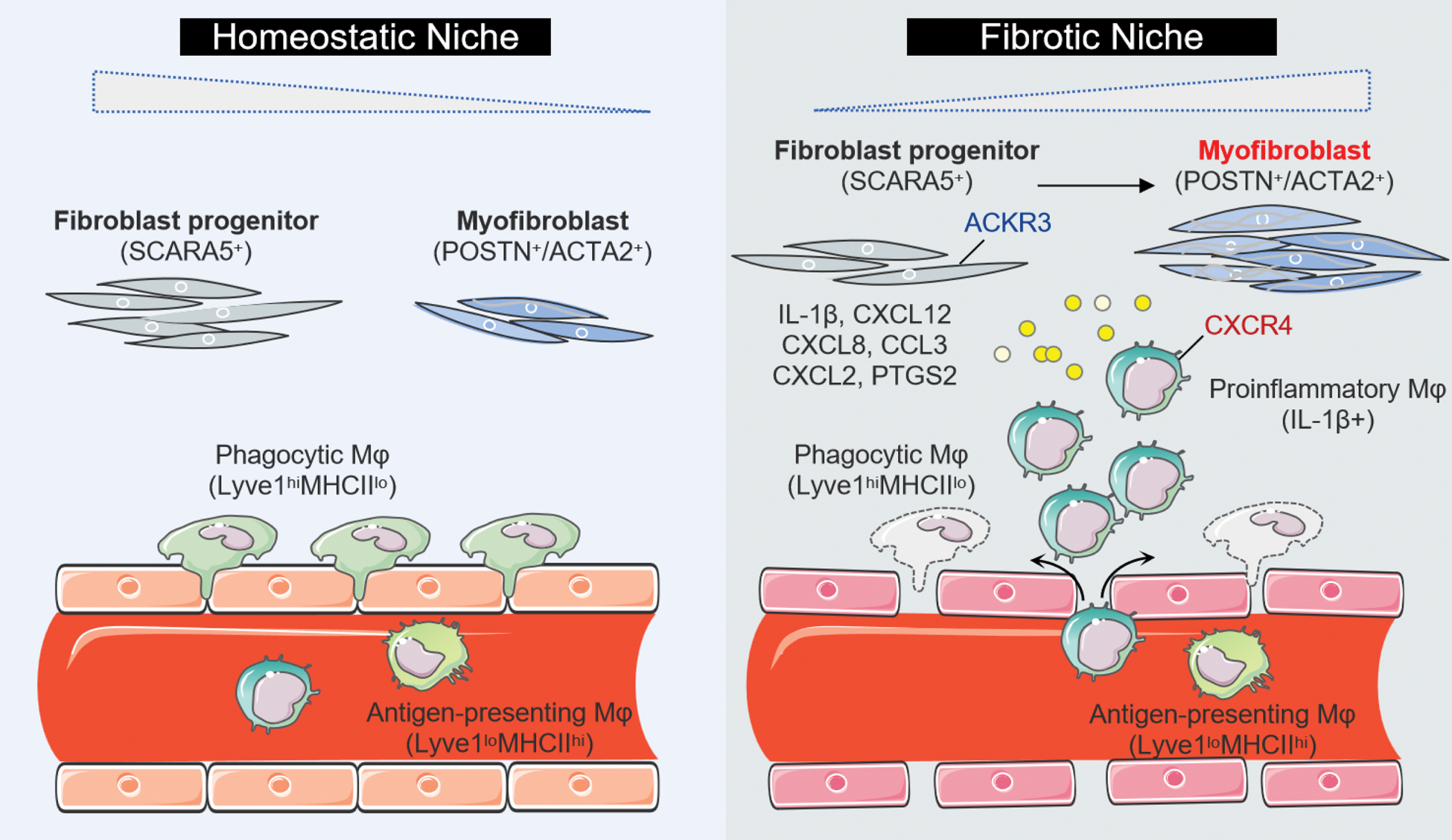
Results: The integration of transcriptomics and tissue structure revealed a fibrotic niche, enriched with fibroblasts and macrophages, which was significantly expanded in SSc and correlated with clinical outcome. To elucidate the specific cell types engaged in spatially educated cell-cell communication, we performed re-clustering and identified an imbalance in fibroblast subsets in SSc, characterized by a decrease in SCARA5 + fibroblast progenitors and an increase in POSTN + / ACTA2 + terminally differentiated myofibroblasts. Pseudotime trajectory analysis indicated divergent differentiation pathways from SCARA5 + fibroblasts to two myofibroblasts, which aligns with observations across different time points in SSc and wound healing models. We also identified three macrophage (Mφ) populations with distinct transcriptional profile in SSc skin versus controls: increased inflammatory Mφ ( IL-1β + ), decreased phagocytic Mφ ( Lyve1 hi MHCII lo ), but unaltered antigen-presenting Mφ ( Lyve1 lo MHCII hi ). We further discovered enhanced interactions between fibroblasts and IL-1β + inflammatory Mφ, suggesting spatial dependency. We identified specific expression of ACKR3 in fibroblast progenitors that diminishes over SSc progression, which may serve to regulate CXCL12/CXCR4-mediated Mφ recruitment and tissue fibrotic remodeling.
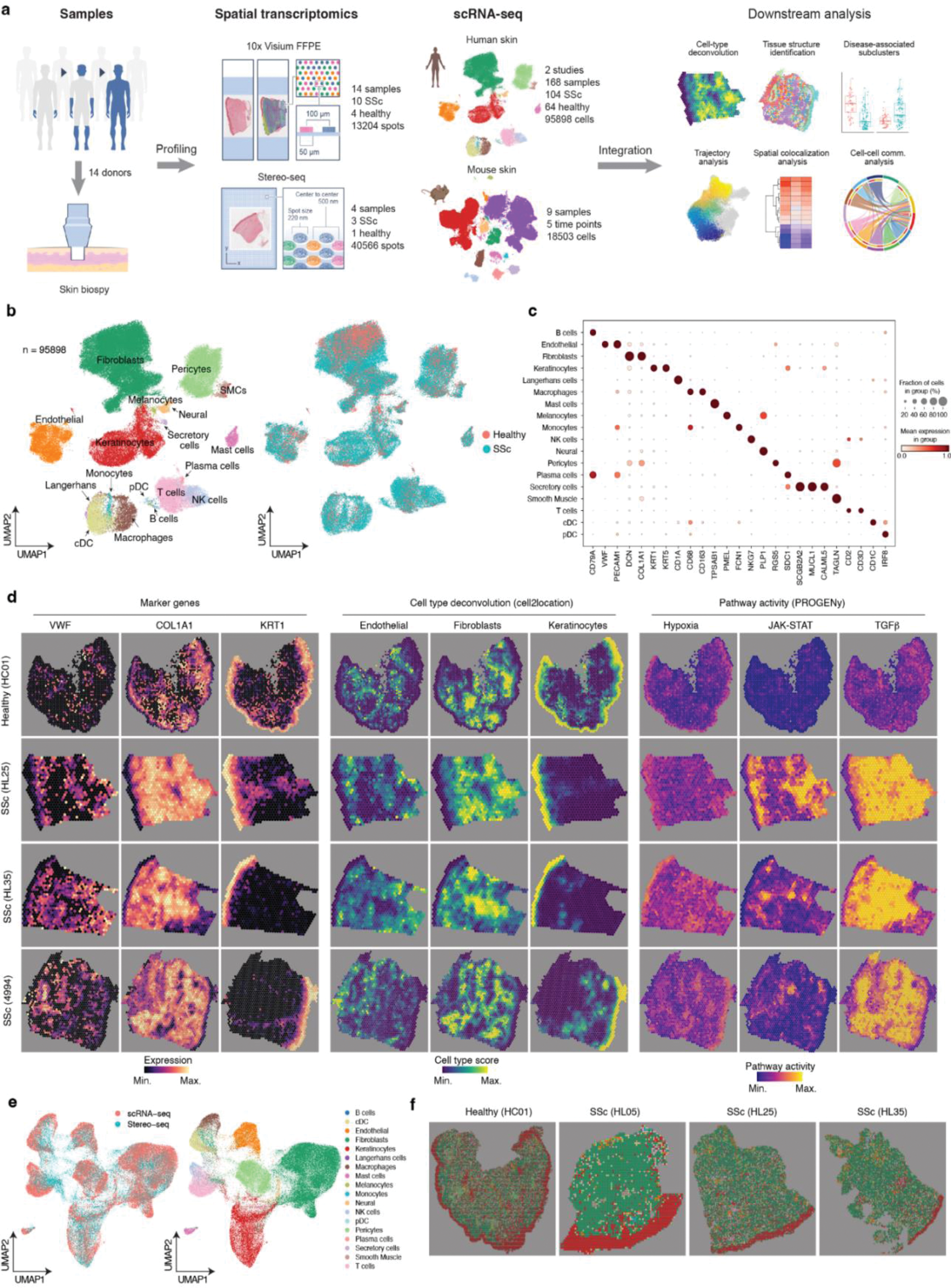
Spatial profiling of human SSc skin. a, Overview of experimental design and analysis workflow. b, UMAP showing integrated scRNA-seq datasets as colored by cell types (left) and conditions (right). SSc skin n=104, Healthy skin n=64. c, Dotplot showing gene expression for marker genes. The colors refer to the mean expression of the genes and the dot sizes represent the proportion of cells expressing the marker genes. d, Visualization in spatial space based on 10× Visium. SSc skin n=10, Healthy skin n=4. Left: vWF, COL1A1, and KRT1 gene expressions in spatial transcriptomics data. Middle: the representative cell types estimated by cell2location. Right: pathway activity in spatial space. e, Integration of scRNA-seq and Stereo-seq data colored by protocols (left) and cell types (right). f, Visualization of cell types in spatial space based on stereo-seq data. SSc skin n=3, Healthy skin n=1. The experiments were performed on adjacent tissue sections to those used for 10× Visium.
A Data-Driven Model of SSc.
Schematic summary illustrates the proposed cellular interplay in SSc skin, which leads to a shift from tissue homeostatic to a fibrogenic environment. The illustration was created by using images from Servier Medical Art
(
Conclusion: We provided an in-depth description at cellular and spatial level of fine-tune regulatory events occurring in SSc, offering spatiotemporal insights and potentially paving the way for advanced mechanistic and therapeutic studies of SSc.
REFERENCES: [1] Distler, J. H. W. et al. Shared and distinct mechanisms of fibrosis. Nat. Rev. Rheumatol. 15, 705–730 (2019).
[2] Alexanian, M. et al. Chromatin remodelling drives immune cell-fibroblast communication in heart failure. Nature 635, 434–443 (2024).
[3] Amrute, J. M. et al. Targeting immune-fibroblast cell communication in heart failure. Nature 635, 423–433 (2024).
[4] Mayr, C. H. et al. Spatial transcriptomic characterization of pathologic niches in IPF. Sci. Adv. 10, eadl5473 (2024).
[5] Del Galdo F, et al. EULAR recommendations for the treatment of systemic sclerosis: 2023 update. Ann Rheum Dis. ard-2024-226430 (2024).
[6] Sinha, S. et al. Fibroblast inflammatory priming determines regenerative versus fibrotic skin repair in reindeer. Cell 185, 4717–4736.e25 (2022).
[7] Chakarov, S. et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 363, eaau0964 (2019).
[8] Tabib, T. et al. Myofibroblast transcriptome indicates SFRP2hi fibroblast progenitors in systemic sclerosis skin. Nat. Commun. 12, 4384 (2021).
[9] Gur, C. et al. LGR5 expressing skin fibroblasts define a major cellular hub perturbed in scleroderma. Cell 185, 1373–1388.e20 (2022).
[10] Rius Rigau, A. et al. Imaging mass cytometry-based characterisation of fibroblast subsets and their cellular niches in systemic sclerosis. Ann. Rheum. Dis. (2024) doi:10.1136/ard-2024-226336.
[11] Rius Rigau, A. et al. Characterization of vascular niche in systemic sclerosis by spatial proteomics. Circ. Res. 134, 875–891 (2024).
[12] Kuppe, C. et al. Decoding myofibroblast origins in human kidney fibrosis. Nature 589, 281–286 (2021).
[13] Kuppe, C. et al. Spatial multi-omic map of human myocardial infarction. Nature 608, 766–777 (2022).
[14] Li, Z. et al. Chromatin-accessibility estimation from single-cell ATAC-seq data with scOpen. Nat. Commun. 12, 6386 (2021).
Acknowledgements: NIL.
Disclosure of Interests: Minrui Liang: None declared, Zhijian Li: None declared, Aleix Rius Rigau: None declared, Wenjie Xie: None declared, Yi-Nan Li: None declared, Alexandru-Emil Matei: None declared, Hejian Zou: None declared, Rui He: None declared, Jörg Distler JHWD is CEO of 4D Science and scientific lead of FibroCure, JHWD has consultancy relationships with Active Biotech, Anamar, ARXX, AstraZeneca, Bayer Pharma, Boehringer Ingelheim, Callidatas, Celgene, Galapagos, GSK, Inventiva, Janssen, Kyverna, Novartis, Pfizer, Quell Therapeutics and UCB, JHWD has received research funding from Anamar, ARXX, BMS, Bayer Pharma, Boehringer Ingelheim, Cantargia, Celgene, CSL Behring, Exo Therapeutics, Galapagos, GSK, Incyte, Inventiva, Kiniksa, Kyverna, Lassen Therapeutics, Mestag, Sanofi-Aventis, RedX, UCB and ZenasBio.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (