

Background: Clinical and biological studies on the Very Early Diagnosis of Systemic Sclerosis (VEDOSS) population have established that very early SSc is a tractable disease state that can offer a window of opportunity for early detection and intervention. In this context, analysis of biosamples from VEDOSS skin biopsies are invaluable for unravelling the very early stages of SSc pathogenesis.
Objectives: Here we aimed to characterise the skin proteome of the distinctive stages of SSc, to identify critical pathways for early detection of biological activity and progression to skin fibrosis.
Methods: SomaScan proteomic analysis (profiling >7000 proteins) was performed on whole skin biopsies from healthy, VEDOSS and SSc patients. Differential expression analysis identified deregulated proteins in the skin, principal component analysis was used to stratify patients, and pathway analysis was used to investigate critical networks responsible for each stage. Pearson correlation coefficients were calculated for clinical data.
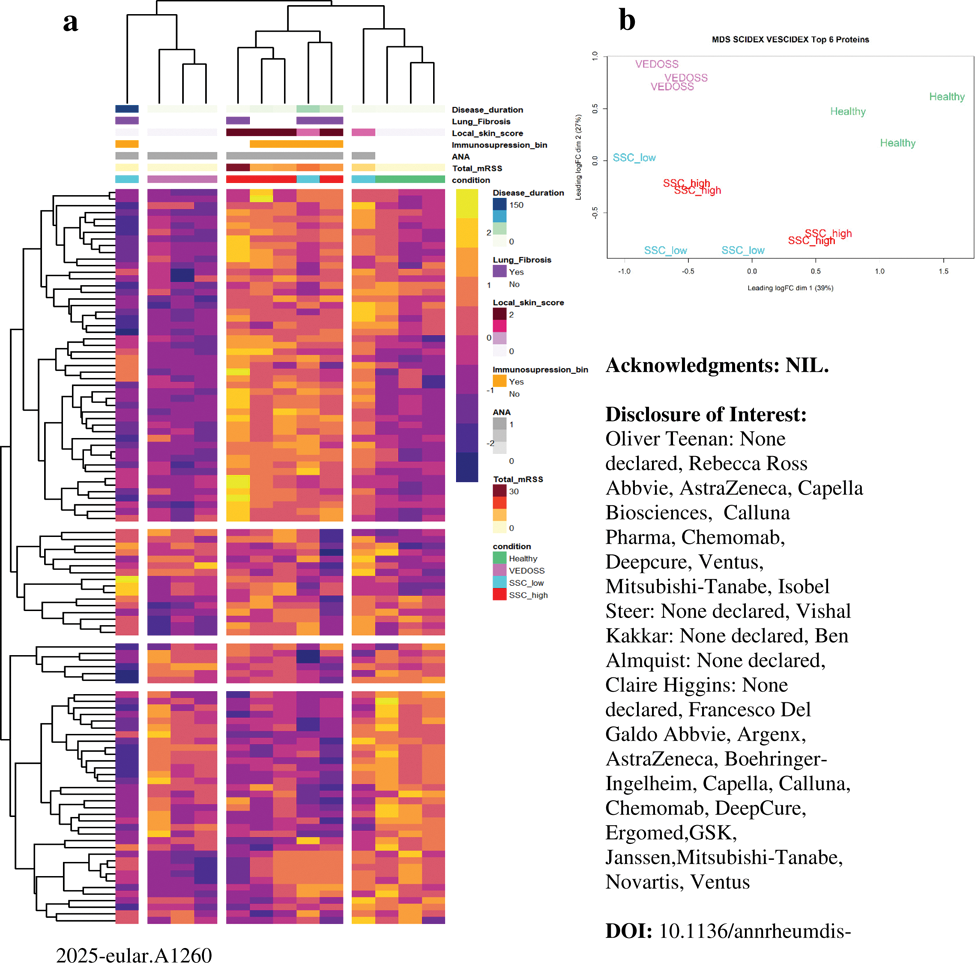
Results: 13 skin biopsies (N=3 Healthy, N=3 VEDOSS, N=7 SSc) were analysed in this study. Unsupervised clustering showed a strong phenotype between groupings of patients. Healthy skin samples clustered together, while SSc biopsies from sites with local modified Rodnan Skin Scores (mRSS) ≥2 showed consistent clustering away from healthy skin. Biopsies from sites with local mRSS <2 showed the least consistent clustering. VEDOSS patients clustered separately despite having no clinical skin involvement, suggesting pre-clinical underlying changes within the skin (Figure 1A). Pearson correlation analysis was performed to identify biomarkers of skin thickness. Proteins with a high degree of correlation with total mRSS, included CCL18 (R2 0.92, P<0.5x10-6), TIMP1 (R2 0.85, P<0.5x10-5), SERPINE2 (R2 0.94, P<0.5x10-7). Local skin score of sampled skin showed high positive correlation with TPSAB1 (R2 0.88, P<0.005) and inverse correlation with CHI3L1 (R2 0.87, P<0.005). Comparative analysis identified an intrinsic proteomic signature of SSc consisting of 27 proteins deregulated at VEDOSS stages which remain deregulated in SSc compared to healthy. Among these, top deregulated proteins include reduction in the amount of CFD (1.8fold, p<0.001), MATN4 (5.9 fold, p <0.5x10-4), PCOLCE2 (1.9 fold change, p <0.005) as well as increased concentration of COMP (2.8-fold increase, p <0.05). The same analysis also revealed a specific VEDOSS signature that is lost with skin fibrosis progression which includes significant loss of proteins involved in cell-adhesion, extracellular matrix organisation and coagulation as well as a decrease in complement factors, whereas metabolic and catabolic gene-ontology sets were overexpressed. Principal Component Analysis (PCA) was employed to identify the minimum number of proteins required to categorise the skin proteome between healthy, clinically affected skin and very early disease without clinically detectable skin fibrosis. While the top 20 coefficients were able to establish a strong stratification of samples, we were able to reduce the number of proteins down to 6 (MATN4, HBB, COMP, CAPS, CD36 and FABP9) through an iterative process which enabled clear independent groupings of all samples by PCA (Figure 1B). MATN4 loss was a consistent biomarker of all stages of SSc including VEDOSS. Overexpression of COMP was a marker of SSc skin but showed limited change at very early stages. On the contrary, CD36 and HBB loss are both markers of VEDOSS stages, returning towards baseline levels in SSc skin. Interestingly, FABP9 overexpression in skin is a marker of early stages of the disease and consistent with less fibrotic skin, but not in advanced SSc skin fibrosis.
Conclusion: Novel investigation of the distinct skin proteome in SSc has identified both classic biomarkers of skin fibrosis and highlighted the specific biology of the skin at the very early stages. Established biomarkers of skin stiffness were identified, with increased levels of TIMP1, CCL18 and SERPINE2. While loss of CFD, MATN4 and PCOLCE2 and the overexpression of ERAP1 has been identified as very early changes in SSc skin remaining consistent as patients progress. Novel insights into the VEDOSS skin proteome and gene ontology identified potentially targetable pathways critical in the establishment and progression of the disease which may highlight new therapeutic intervention strategies at very early stages of the disease.
A. Top significantly deregulated proteins, p<0.01, logFC > -/+ 0.585. Comparisons included VEDOSS (pink) vs healthy (green), SSc high (red) vs VEDOSS, SSc high vs healthy, SSc low (blue) vs healthy. Further metadata is coloured above. Data shows Z scores, unsupervised clustering of samples and proteins. Samples and proteins were allowed to cut into 4 by k-means clustering to represent the 4 groups input. B. PCA plot, representing the SCIDEX and VESCIDEX, calculated from 6 genes. Samples are coloured by disease category.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: Oliver Teenan: None declared, Rebecca Ross Abbvie, AstraZeneca, Capella Biosciences, Calluna Pharma, Chemomab, Deepcure, Ventus, Mitsubishi-Tanabe, Isobel Steer: None declared, Vishal Kakkar: None declared, Ben Almquist: None declared, Claire Higgins: None declared, Francesco Del Galdo Abbvie, Argenx, AstraZeneca, Boehringer-Ingelheim, Capella, Calluna, Chemomab, DeepCure, Ergomed,GSK, Janssen,Mitsubishi-Tanabe, Novartis, Ventus.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (