

Background: Anti-neutrophil cytoplasmic autoantibody (ANCA)-associated vasculitis (AAV) is a rare and severe autoimmune disease, characterized by ANCAs targeting myeloperoxidase (MPO) or (PR3). ANCA-specific B cells have a central role in the pathophysiology of this disease, however current knowledge of their specific phenotypic markers and functional pathways is limited. Rituximab is an effective treatment, but relapses of the disease remain a challenge. Therefore, there is an unmet need for novel therapeutic approaches that achieve durable remission.
Objectives: The aim of this study is to identify and characterize ANCA-specific B cells in AAV patients to gain insight into the pathogenesis of AAV and identify novel targets for therapeutic development.
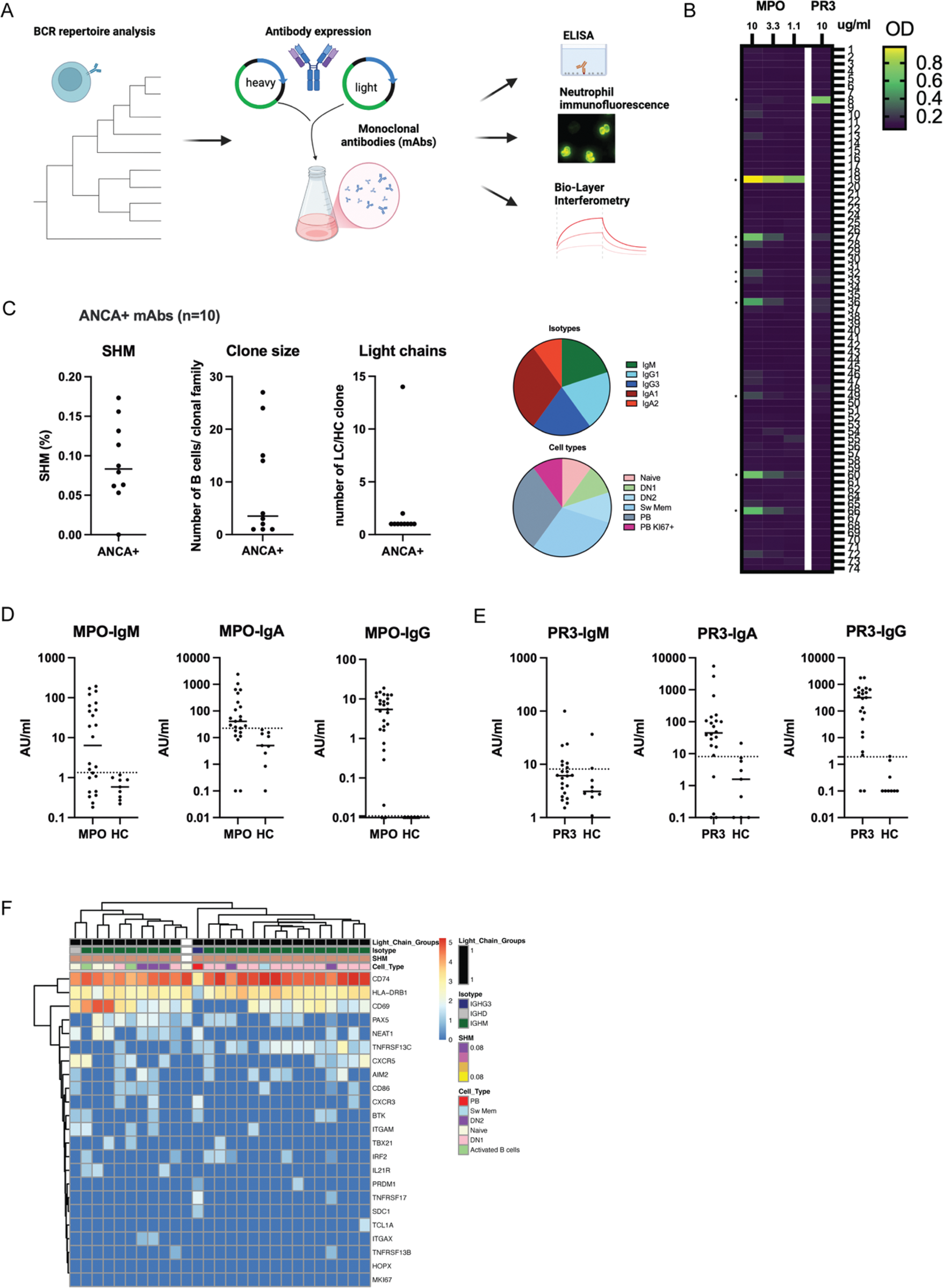
Methods: B cells were isolated by negative selection from PBMCs of 8 anti-MPO + , 8 anti-PR3 + AAV patients and 5 healthy controls (HCs). BCR repertoires and whole transcriptomes of B cells were analyzed with scRNA sequencing combined with CITE-seq (Figure 1A). Bioinformatic analyses were performed to study transcriptional pathways of B cells and their clonal families. Monoclonal antibodies (n=74) were expressed of highly expanded and/or mutated B cells and their target antigens were screened with ELISAs (Figure 2A). ANCA-IgG, -IgM and IgA antibodies were measured in serum of 24 anti-MPO, 23 anti-PR3 AAV patients and 9 HCs.
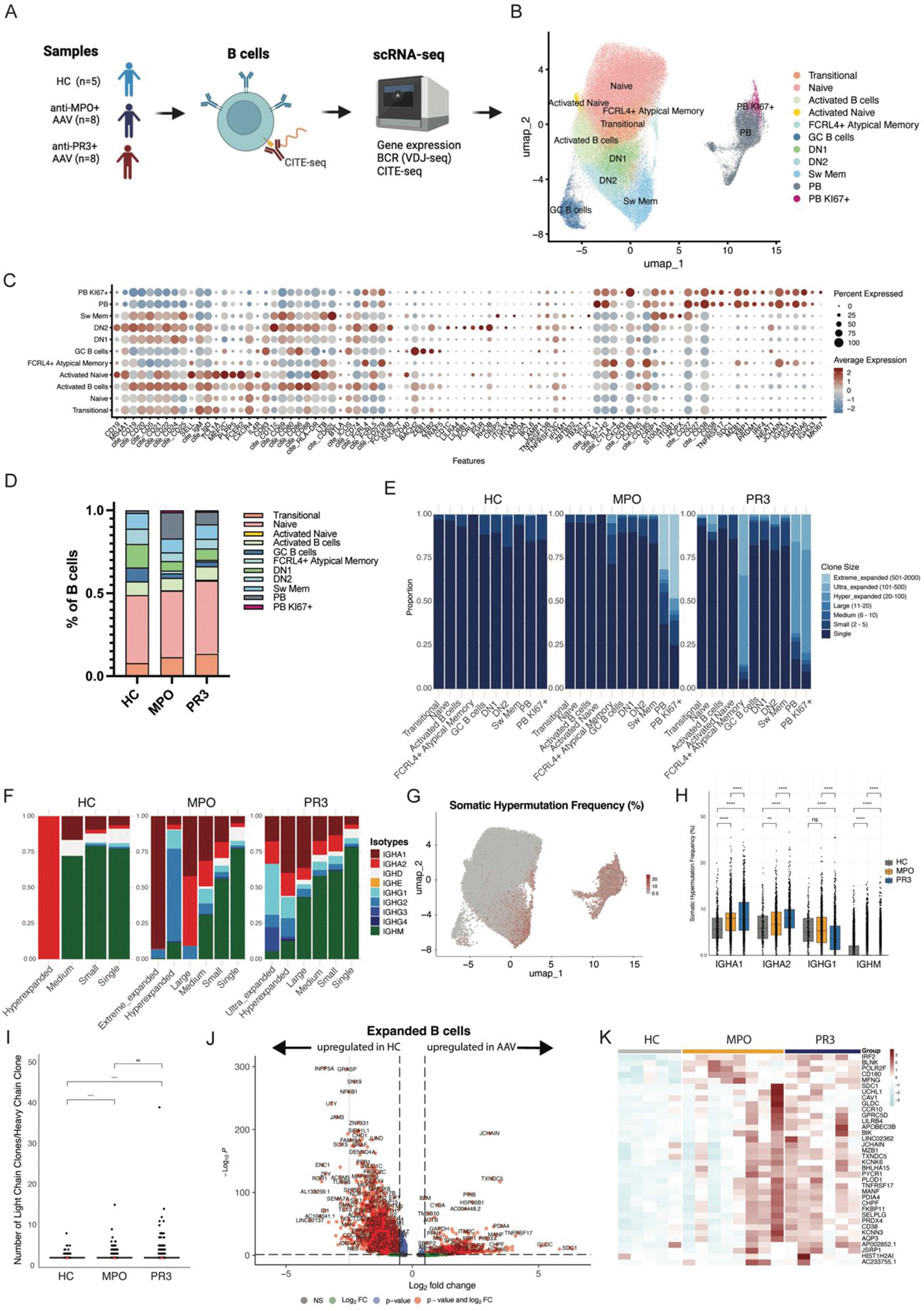
Results: B cell subsets were identified after unsupervised clustering and annotated based upon gene and CITE-seq expression, resulting in transitional, naive, activated, double negative (DN)1, DN2, (atypical) memory B cell and plasmablast (PB) subsets (Figure 1B-C). Both anti-MPO + - and PR3 + AAV patients had significantly decreased numbers of DN1 B cells as compared to HC, whereas anti-MPO + AAV patients had significantly increased PBs (Figure 1D). Both groups of AAV patients had large populations of clonally expanded B cells, predominantly in the PB compartment (Figure 1E), while specifically anti-PR3 + AAV patients had expansions in the FcRL4 + atypical memory B cells. The isotypes of the expanded B cells were predominantly IgA1-2, and IgG1-2 in AAV, in contrast to HC (Figure 1F). Additionally, the somatic hypermutation (SHM) frequency (Figure 1G) was significantly higher in naive, transitional, DN2 (only anti-MPO) and switched memory B cells in both AAV patients versus HC, while it was lower in DN1 (data not shown). Also, SHM was increased for IgA1, IgA2, IgG2 (only anti-PR3 + ), while it was lower for IgM and IgG1 (only anti-PR3 + ) as compared to HCs (Figure 1H). Analyses of light chains demonstrated that both anti-MPO + and anti-PR3 + AAV patients had a higher number of light chain rearrangements within the heavy chain based clonal families (Figure 1I), and overall the lambda usage was significantly higher in anti-MPO + (37.7%) and anti-PR3 + (36.0%) compared to HC (30.8%, p<0.0001). Differential gene expression analysis of the expanded B cell clones identified several significant differentially expressed genes in AAV as compared to HC, including TNFRSF17 , SDC1 and JCHAIN (Figure 1J-K), which are involved in antibody production, B cell survival and migration. BCRs of highly expanded and/or mutated B cells were selected for monoclonal antibody (mAb) production (Figure 2A). Using MPO- and PR3-ANCA assays, we identified 9 mAbs that bound MPO and 1 PR3, within 5 different patients (*, Figure 2B). The ANCA + mAbs had varying SHM of 0 – 17%, clone sizes 1-27, and 2/10 had light chain rearrangements (Figure 2C). Isotypes were IgM, IgA1/2, IgG1/3, while varying B cell types were present (Figure 2C). Importantly, serum autoantibody analyses of these patients together with an additional AAV cohort demonstrated that MPO-IgG, IgM and IgA autoantibodies were present in anti-MPO + (Figure 2D), while in anti-PR3 + patients we found mostly PR3-IgA and PR3-IgG, and only in a small subset of patients PR3-IgM autoantibodies (Figure 2E). The characterization of one large expanded clonal family (mAb 19), which shared the exact same BCR, demonstrated that varying isotypes (IgM, IgD, IgG3) and cell types (naive, DN1, DN2, Sw Mem, PB) were present, but with changes in CD69 , CXCR5 and TNFRSF13 expression (Figure 2F), suggesting a recent activation.
Conclusion: In conclusion, our single cell RNA-seq analysis of B cell subsets in AAV patients reveals significant alterations in B cell populations and their functional characteristics compared to HCs. AAV patients exhibit clonal expansions, particularly in the plasmablast compartment, alongside increased somatic hypermutation rates in various B cell subsets and IgA isotypes. Our findings suggest a potential ongoing mucosal immune activation in these AAV patients. Our characterization of ANCA-specific B cells and their dysregulated activation provides insight into the pathogenic mechanisms underlying AAV and underscores the potential for future therapeutic applications and biomarker development.
Overview of scRNA sequencing of B cells in AAV.
Overview of monoclonal antibody (mAb) analyses.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (