

Background: Sjögren’s disease (SjD) is a complex autoimmune disorder characterized by a significant heterogeneity among patients. Recently, three subsets of patients have been identified through cluster analysis: 1) B-cell active disease with low symptom burden (BALS), 2) high systemic disease activity (HSA), and 3) low systemic disease activity with high symptom burden (LSAHS) [1,2]. The pathophysiological pathways underlying these distinct clusters remain to be elucidated.
Objectives: To assess the blood transcriptomic signature of SjD patients according to their cluster. Specifically, to investigate the BALS subgroup, characterize its transcriptomic profile and identify pathways associated with the disease progression.
Methods: Differential expression analysis (DEA) was performed using transcriptomic data from the ASSESS cohort (GSE140161), a French multicentric study including 351 blood samples from SjD patients. Samples were classified into subgroups based on criteria established by Nguyen et al. [1]: BALS (n=165), HSA (n=142), and LSAHS (n=44). Within the BALS group, further stratification was conducted to define an evolutive subgroup requiring new immunosuppressive therapy or developing lymphoma during follow-up (BALS_IS_Ly, n=15) and a reference group (BALS_Ref, n=150), as described by Nguyen et al. [2]. Pairwise comparisons were carried out across all subgroups, followed by pathway enrichment analysis using Gene Set Enrichment Analysis (GSEA) to identify relevant biological pathways.
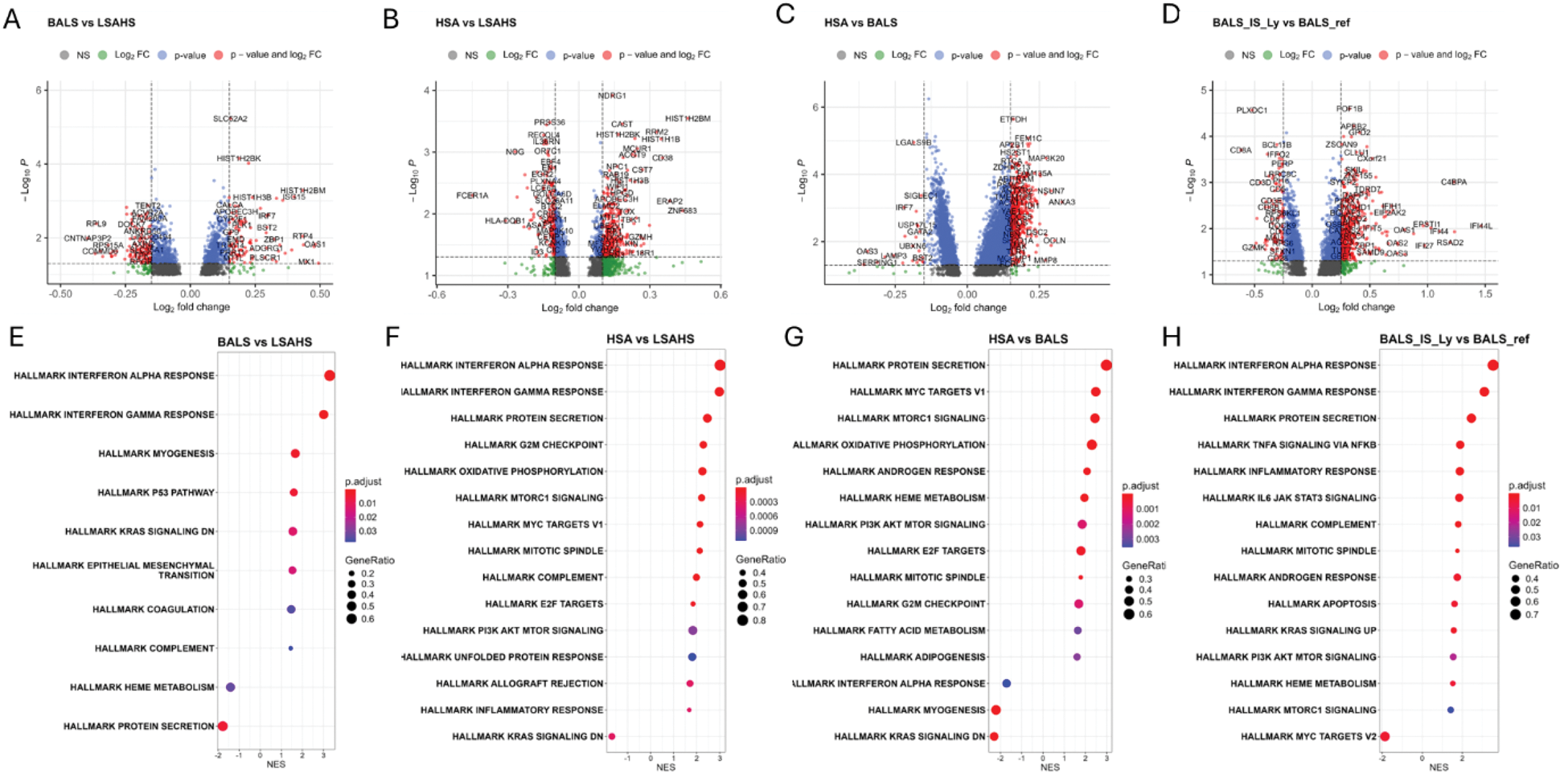
Results: DEA resulted in 1,073 DEGs between the BALS and LSAHS groups, 906 DEGs between the HSA and LSAHS groups, and 5,168 DEGs between the HSA and BALS groups (Figures 1A-C). Enrichment analyses highlighted strong interferon signatures for both BALS and HSA groups compared to LSAHS, with normalized enrichment scores (NES), a metric indicating the power of up-downregulation of a pathway, approaching 3 in both cases (Figures 1E-F). Given that the BALS cluster may represent an earlier stage with a higher risk of progression [1], it is crucial to identify the mechanisms driving this progression. To address this, we first conducted a comparative analysis of HSA versus BALS. The analysis revealed the activation of several pro-inflammatory pathways in the HSA cluster, including MYC, mTOR, and oxidative phosphorylation. The loss of regulatory pathways might be as a key mechanism leading to the transition from BALS to HSA. Interestingly, a downregulation of the interferon (IFN) signature and of the KRAS pathway was observed in the HSA cluster compared to the BALS cluster (Figures 1G). The KRAS pathway is well known in oncology, where it has been shown to induce detrimental immune modulation in the context of anti-cancer surveillance while playing a protective role in autoimmune contexts. Additionally, a comparison of BALS_IS_LY versus BALS_Ref identified 1,891 DEGs (Figure 1D), with a strong enrichment for IFN signatures (Figure 1H). These findings suggest that IFN signaling pathways may mediate the progression of BALS toward a more active disease phenotype.
Conclusion: This study identifies a strong IFN signature in the two most active disease groups (HSA and BALS) compared to the low systemic activity group (LSAHS) [1]. In patients of the BALS cluster, IFN might support the progression toward a more active disease. In a 2nd step activation of pro-inflammatory and cellular proliferation signature might replace the IFN signature and support the evolution toward a high systemic presentation. These findings provide valuable insights into the immunological transitions linked to disease progression in SjD patients, which may inform the development of personalized therapeutic strategies.
REFERENCES: [1] Nguyen Y, Nocturne G, Henry J, Ng WF, Belkhir R, Desmoulins F, et al. Identification of distinct subgroups of Sjögren’s disease by cluster analysis based on clinical and biological manifestations: data from the cross-sectional Paris-Saclay and the prospective ASSESS cohorts. Lancet Rheumatol. 1 avr 2024;6(4):e216‑25.
[2] Nguyen Y, Beydon M, Gottenberg JE, Morel J, Perdriger A, Dernis E, et al. Distinct pathophysiological pathways support stratification of Sjögren’s disease based on symptoms, clinical, and routine biological data. Arthritis Rheumatol Hoboken NJ. 25 déc 2024.
Transcriptomic signatures for subgroups of Sjogren patients. (A–D) Volcano plots depicting DEGs for (A) BALS vs. LSAHS, (B) HSA vs. LSAHS, (C) HSA vs. BALS and (D) BALS_IS_Ly vs. BALS_ref. The log fold-change (LogFC) cutoff is set at 0.15, with a p-value threshold of < 0.05. (E–H) Dotplots representing GSEA results for (E) BALS vs. LSAHS, (F) HSA vs. LSAHS, (G) HSA vs. BALS and (H) BALS_IS_Ly vs. BALS_ref comparisons.
Acknowledgements: The authors sincerely thank all the patients for their participation and the physicians who contributed to the inclusion of patients in the ASSESS cohort. They also wish to extend their gratitude to all the principal investigators involved in the NECESSITY consortium.
Disclosure of Interests: Sacha E Silva-Saffar: None declared, Yann Nguyen: None declared, Raphaèle Seror GSK, BMS, Boehringer Ingelheim, Janssen Pharmaceuticals, Jacques-Eric Gottenberg BMS, Pfizer, Lilly, Abbvie, AstraZeneca, Gilead, Galapagos, MSD, Roche-Chugai, Sanofi, UCB, Xavier Mariette Sanofi, Servier, BMS, Anna Niarakis Sanofi, Gaetane Nocturne Amgen, Novartis.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (