

Background: In active rheumatoid arthritis (RA), a macrophage-rich lining-layer was shown to be populated with inflammatory dendritic cells (iDC3) that promote synovitis [1]. However, the stromal compartment which may drive iDCs niche formation was not identified.
Objectives: To identify the interaction of synovial fibroblasts with iDC3 in RA and to compare with other joint diseases.
Methods: Synovial tissue was obtained by ultrasound-guided synovial biopsy from inflamed joints of 15 patients with RA, 12 patients with PsA, five patients with undifferentiated arthritis (UA), and five patients with osteoarthritis (OA). Six synovial tissues of patients with traumatic injuries without inflammatory arthritis were used as controls (TRA). Synovial biopsies were digested and used for single-cell RNA sequencing (scRNAseq). PBMCs of additional five patients with RA and six patients with PsA were also used for scRNAseq and merged with synovial cells. 217’109 cells (180’473 synovial and 36’636 blood cells) were used for scRNA-seq analysis (mean of 4’197 synovial and 3’330 blood cells after QC per patient). PBMC scRNAseq data was integrated with the data from the synovium tissue and used only for clustering. Seurat R package was used for scRNAseq data analysis. Cell types were manually annotated and integrated using STACAS. MultiNicheNet and CellChat R packages were used for cell-cell interaction analysis. Four additional synovial biopsies (2 RA, 1 PsA and 1 OA) were paraffin-embedded and used for spatial transcriptomics analysis. STutility R package was used for data preprocessing and neighbourhoods annotation. UCell R package was used to calculate signature values.
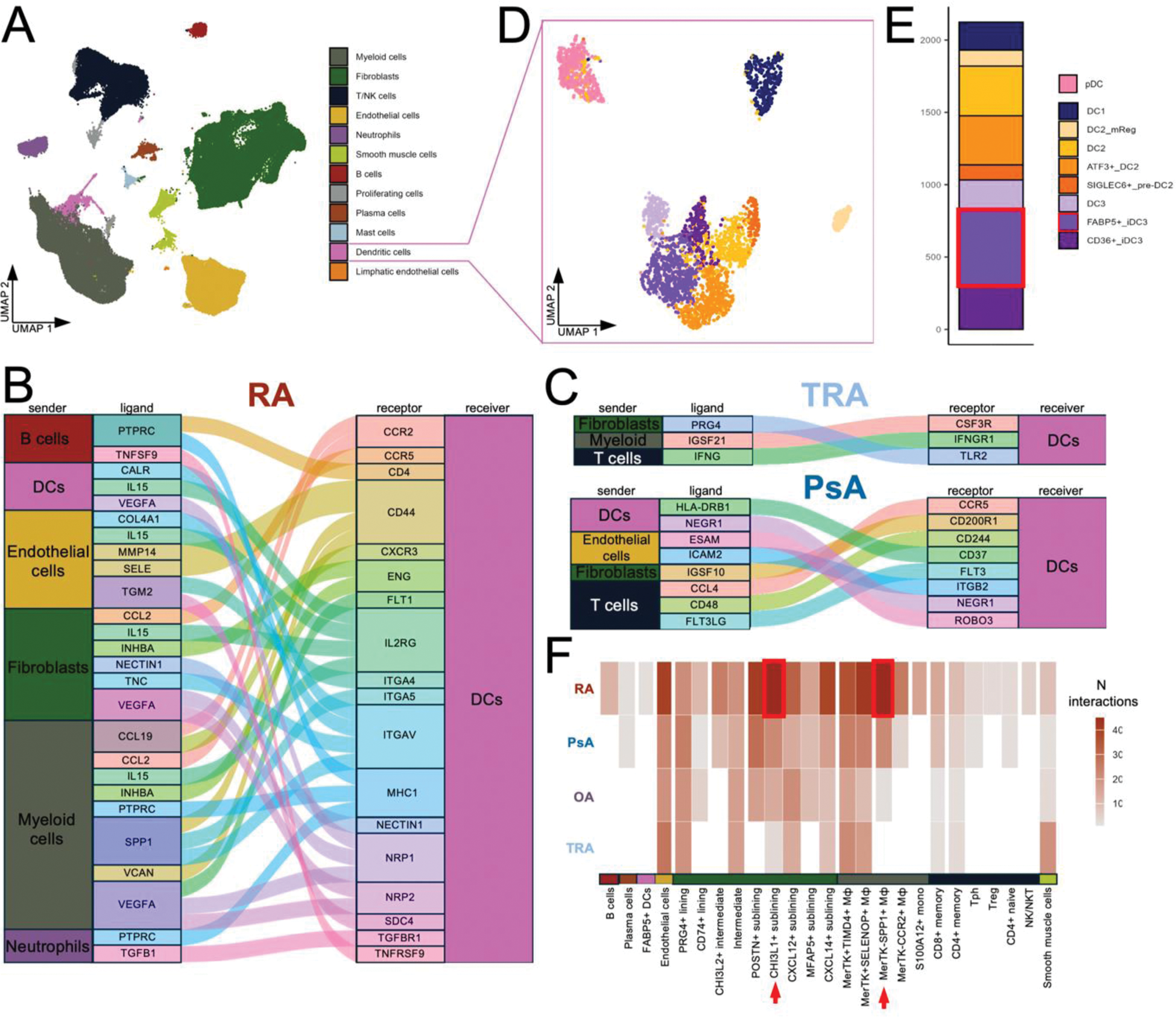
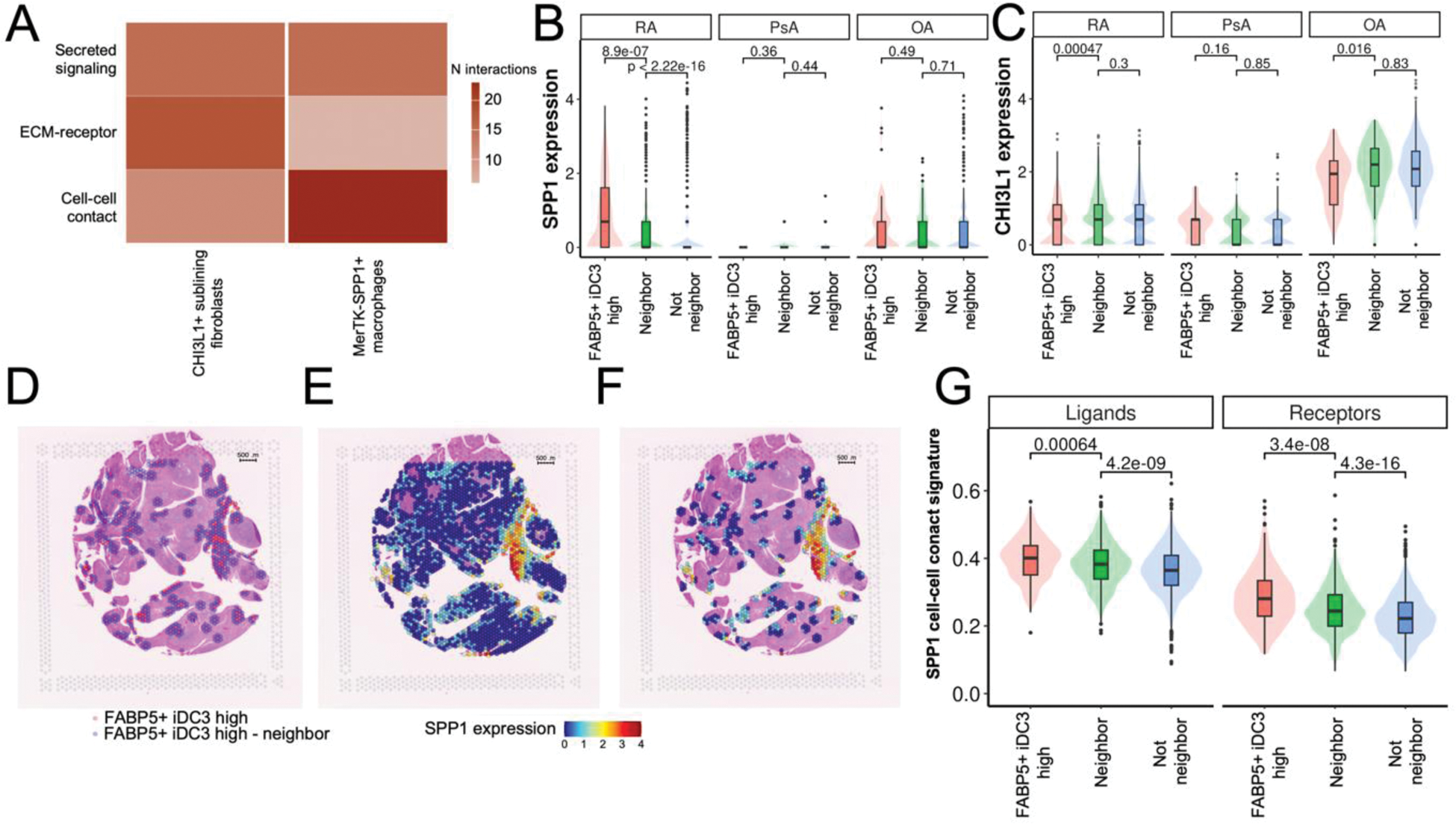
Results: Analysis of scRNAseq data showed that myeloid cells and fibroblasts are the most abundant cell types in the synovium (27% (±6%) and 33% (±16%) of all synovial cells, respectively), while DCs account for only 1.5% (±0.5%) of the synovium (Figure 1A). Cell-cell interactions with DCs were enriched in RA synovium (Figure 1B) in comparison to PsA and controls (Figure1C). Analysis of DC subsets revealed the presence of plasmacytoid DCs and eight DC subpopulations, including FABP5+ inflammatory DC3 (FABP5+ iDC3) (Figure1D). This subpopulation was shown to receive the most interactions among DCs according to the CellChat analysis (Figure 1E). Most of these interactions was coming from fibroblasts, specifically from the CHI3L1+ fibroblasts and MerTK-SPP1+ macrophages (Figure 1F). We previously showed that CHI3L1+ fibroblasts are more abundant in RA synovium in comparison to PsA, OA and controls [2]. Analysis of the number of interactions by type of signalling showed that SPP1+ macrophages interacted with FABP5+ iDC3 mostly via cell-cell contact interactions, while CHI3L1+ fibroblasts relied on secreted signalling and ECM-receptor interactions (Figure 2A). To confirm proximity of FABP5+ iDC3 with SPP1+ macrophages we analyzed spatial transcriptomics data. This analysis allowed identification of spots with high expression of FABP5+ iDC3 signature (CLEC10A+CD14+FABP5+). Neighbours to these spots were annotated (Figure 2D) and expression values of SPP1 (a marker for MerTK-SPP1+ macrophages) and CHI3L1 (a marker for CHI3L1+ fibroblasts) were analyzed. In accordance with the high number of predicted cell-cell contact interactions, SPP1 expression was the highest in FABP5+ iDC3 high regions and decreased with distance (Figure 2B, E, F), showing high abundance of SPP1+ macrophages in proximity with FABP5+ iDC3. Notably, this pattern was observed only in RA samples, but not in PsA or OA. Additionally, we analyzed the expression pattern of ligands and receptors (e.g. SPP1-CD44, ITGB2- ICAM1, etc) that were predicted to contribute to cell-cell contact interactions between FABP5+ iDC3 and SPP1+ macrophages. Expression of those ligands and receptors also showed a positive correlation with distance to FABP5+ iDC3 high regions (Figure 2G). At the same time, CHI3L1 expression did not show correlation with the distance to FABP5+ iDC3 high areas (Figure 2C), supporting the observation that CHI3L1+ sublining fibroblasts rely on not cell-cell contact-dependent interactions to communicate with FABP5+ iDC3.
A – UMAP of all synovial cell types. B – Alluvial plot of signalling received by DCs among the top 1000 signalling in RA synovium. C – Alluvial plots of signalling received by DCs among top 1000 signalling in healthy and PsA synovium. D – UMAP of DC subpopulations. E – Bar plot of the number of interactions received per DC subpopulation with probability > 8%. F – Heatmap of the number of interactions received by FABP5+ iDC3 from different cell types across diagnosis.
A – Heatmap of the number of interactions received by FABP5+ iDC3 from CHI3L1+ sublining fibroblasts and MerTK-SPP1+ macrophages by type of signalling. B – Boxplot with violin plots of SPP1 expression values across FABP5+ iDC3 neighbourhoods across diagnosis in spatial transcriptomics data. D – Representative spatial transcriptomics RA slide with FABP5+ iDC3 high spots and their neighbours. E – Spatial transcriptomics RA slide with SPP1 expression values. F – Representative spatial transcriptomics RA slide with SPP1 expression values in FABP5+ iDC3 high neighbourhoods only. G - Boxplot with violin plots of SPP1 cell-cell contact ligands and receptors signature values, respectively, across FABP5+ iDC3 high neighbourhoods.
Conclusion: Our data suggest close interactions between FABP5+ inflammatory DCs, CHI3L1+ fibroblasts and SPP1+ macrophages in RA synovium in comparison to PsA, OA and controls. We propose that these three cell types engage in disease-specific cellular interactions, building an inflammatory niche that may drive inflammation.
REFERENCES: [1] MacDonald et al., 2024, Immunity 57, 1–20
[2] Khmelevskaya et al., 2024, Annals of the Rheumatic Diseases 83:590.
Acknowledgements: NIL.
Disclosure of Interests: Alexandra Khmelevskaya: None declared, Miranda Houtman: None declared, Kristina Buerki: None declared, Chantal Pauli: None declared, Oliver Distler Oliver DIstler has/had consultancy relationships with and/or has served as a speaker for the following companies in the area of potential treatments for systemic sclerosis and its complications in the last three calendar years: 4P-Pharma, Abbvie, Acceleron, Acepodia Biotech, Aera, Alcimed, Altavant, Amgen, AnaMar, Anaveon AG, Argenx, AstraZeneca, Blade, Bayer, Boehringer Ingelheim, Calluna (Arxx), Cantargia AB, Catalyze Capital, Corbus, CSL Behring, Galderma, Galapagos, Glenmark, Gossamer, Horizon, Janssen, Kymera, Lupin, Medscape, MSD Merck, Miltenyi Biotec, Mitsubishi Tanabe, Nkarta Inc., Novartis, Orion, Pilan, Prometheus, Quell, Redxpharma, Roivant, EMD Serono, Topadur and UCB, Patent issued “mir-29 for the treatment of systemic sclerosis” (US8247389, EP2331143). Co-founder of CITUS AG, Oliver DIstler has/had consultancy relationships with and/or has served as a speaker for the following companies in the area of potential treatments for systemic sclerosis and its complications in the last three calendar years: 4P-Pharma, Abbvie, Acceleron, Acepodia Biotech, Aera, Alcimed, Altavant, Amgen, AnaMar, Anaveon AG, Argenx, AstraZeneca, Blade, Bayer, Boehringer Ingelheim, Calluna (Arxx), Cantargia AB, Catalyze Capital, Corbus, CSL Behring, Galderma, Galapagos, Glenmark, Gossamer, Horizon, Janssen, Kymera, Lupin, Medscape, MSD Merck, Miltenyi Biotec, Mitsubishi Tanabe, Nkarta Inc., Novartis, Orion, Pilan, Prometheus, Quell, Redxpharma, Roivant, EMD Serono, Topadur and UCB, Research Grants: BI, Kymera, Mitsubishi Tanabe, UCB, Adrian Ciurea: None declared, Andreas Ramming: None declared, Ursula Fearon: None declared, Caroline Ospelt: None declared, Raphael Micheroli: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (