

Background: Rheumatoid arthritis (RA) is a chronic autoimmune inflammatory disease that affects approximately 5 in 1000 adults worldwide, characterized by joint swelling, pain, and progressive joint damage [1–3]. ACPA, a key diagnostic marker for RA, demonstrates high specificity (>90%) but limited sensitivity (30–60%) [4, 5], leaving a significant proportion of patients undiagnosed in the early stages. ACPA-negative RA (ACPA– RA), which constitutes approximately 40% of cases, differs from ACPA-positive RA (ACPA+ RA) in disease progression and treatment response [6, 7]. Previous studies have highlighted subgroup-specific genetic, immune, and microbiome differences [8, 9], but the biomolecular underpinnings in blood remain underexplored. Multi-omics approaches integrating proteomics and metabolomics offer a promising avenue to address this critical knowledge gap.
Objectives: To identify subgroup-specific immune and metabolic signatures in ACPA– and ACPA+ RA using an integrative multi-omics approach; and to evaluate the potential of these biomolecular profiles in distinguishing RA subgroups and controls.
Methods: Figure 1 provides our methodological overview. Plasma samples were collected from 120 participants, comprising 40 ACPA– RA, 40 ACPA+ RA, and 40 healthy controls. Proteomic and metabolomic profiling were performed using the SomaScan Assay and Metabolon Discovery HD4™ platform, respectively. A total of 8,334 biomolecular features (7,273 proteins and 1,061 metabolites) were analyzed. Statistical comparisons, pathway enrichment analyses, and machine learning methods were utilized to identify subgroup-specific biomolecules and construct classification models. The diagnostic performance of the integrative multi-omics approach was evaluated using area under the receiver operating characteristic curve (AUC).
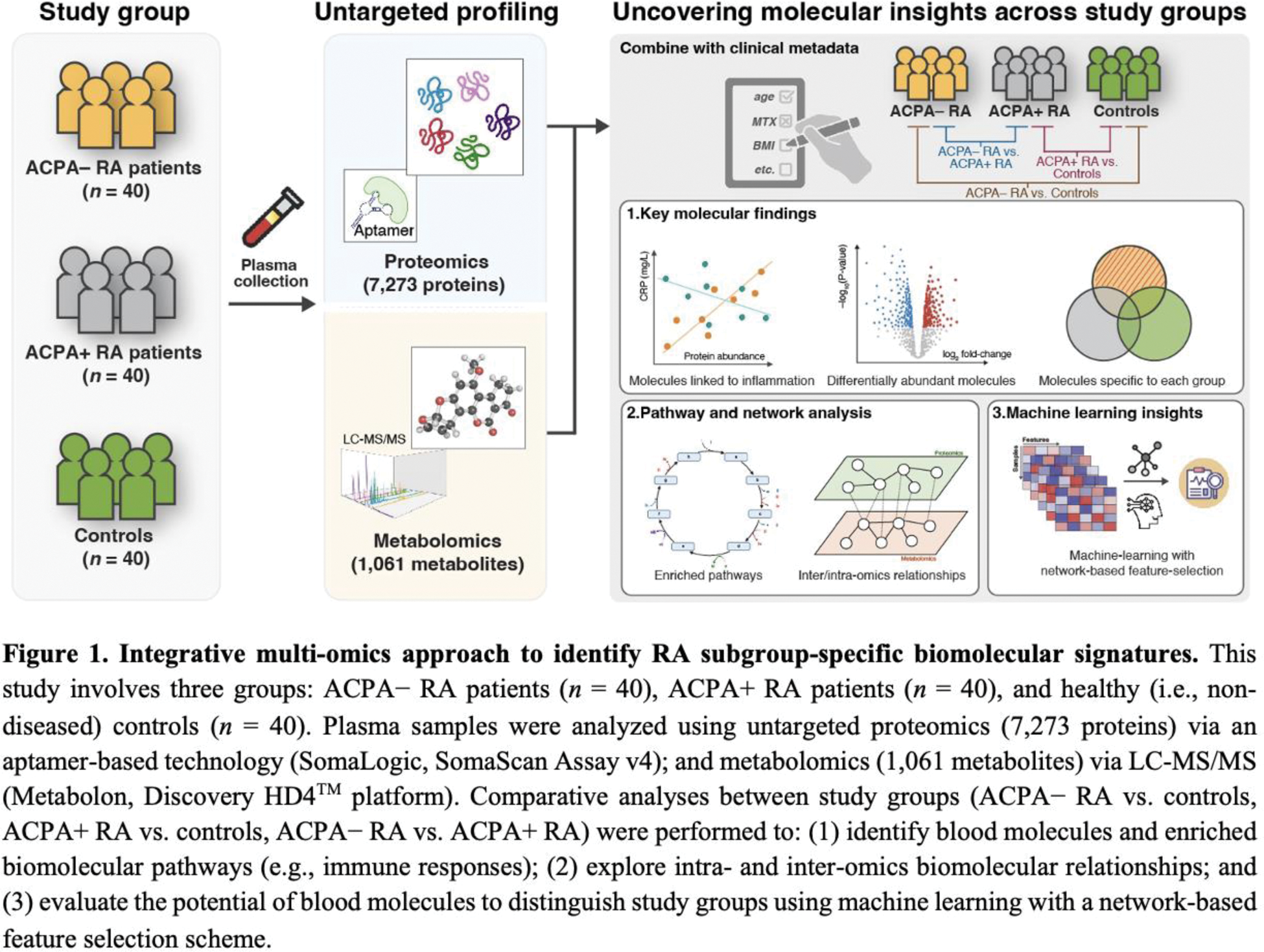
Results: Integrative multi-omics profiling revealed over 100 distinct biomolecules, including cytokines (e.g., CCL7, IL-1Ra, IL-17C; see Figure 2) and metabolites (e.g., pyruvate, lactate, sphingomyelins), that differentiate ACPA– RA, ACPA+ RA, and controls. Lipid metabolic pathways were specifically enriched in ACPA– RA, characterized by elevated fatty acid synthase (FASN) and downstream metabolites. Cytokine profiles demonstrated subgroup-specific patterns, with IL6, CXCL11, and IL-1Ra elevated in ACPA– RA compared to controls. Proteins such as MMP19, and metabolites including biliverdin, exhibited strong subgroup-specific correlations with clinical markers like erythrocyte sedimentation rate and Disease Activity Score 28. Machine learning models integrating proteomic and metabolomic data achieved high diagnostic accuracy, with an AUC of 0.93 for distinguishing ACPA– RA from controls.
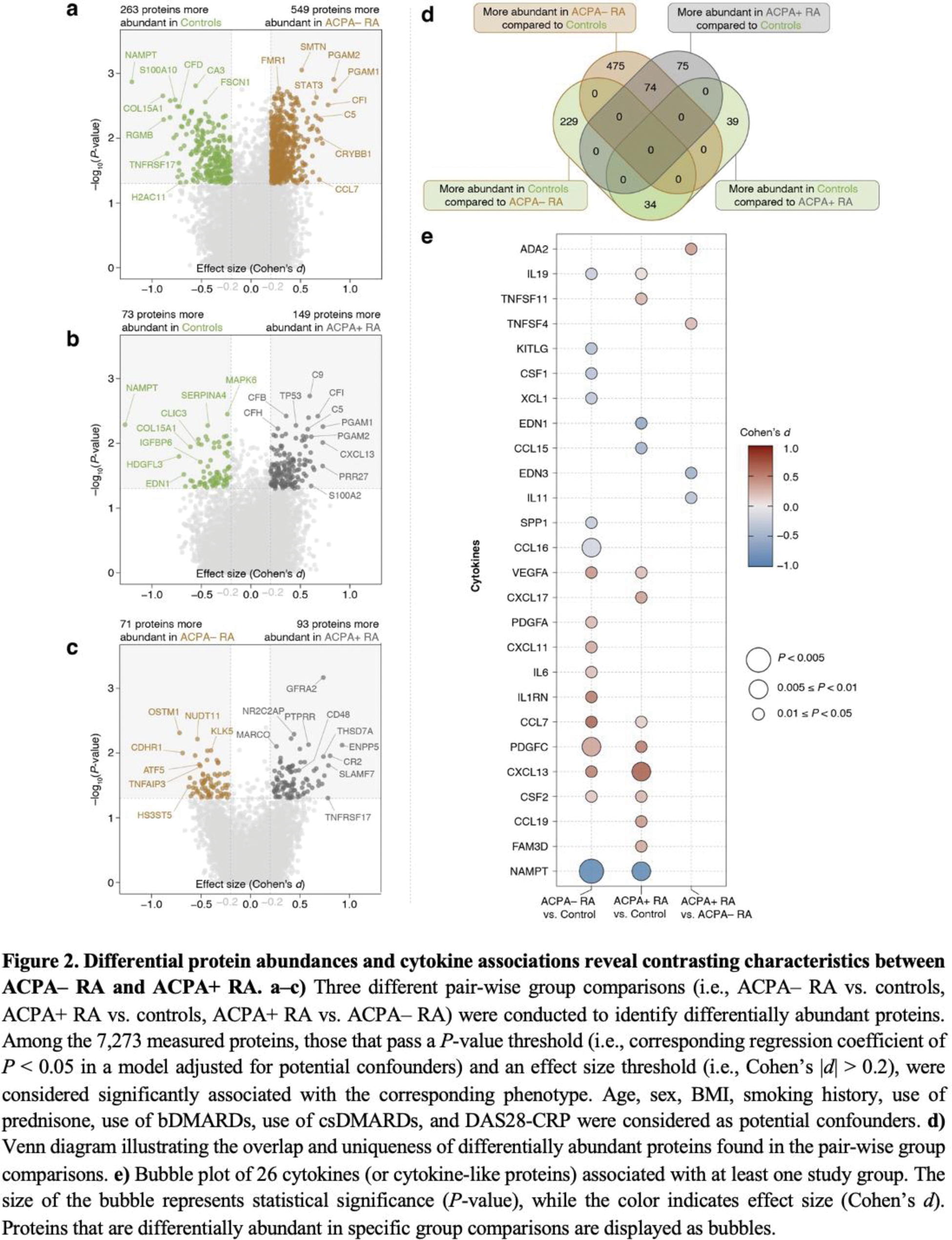
Conclusion: This study presents the first integrative analysis of plasma proteomics and metabolomics in RA subgroups, uncovering distinct immune and metabolic signatures in ACPA– and ACPA+ RA. The findings highlight lipid metabolism dysregulation as a defining feature of ACPA– RA and demonstrate the potential of multi-omics integration for advancing RA diagnostics and precision medicine. Future studies should validate these biomarkers in diverse populations and explore their translational applications in clinical settings.
REFERENCES: [1] D. Aletaha, J. S. Smolen, Diagnosis and management of rheumatoid arthritis: A review. JAMA 320 , 1360–1372 (2018).
[2] J. S. Smolen, D. Aletaha, A. Barton, et al. , Rheumatoid arthritis. Nat. Rev. Dis. Primers 4 , 18001 (2018).
[3] K. Li, M. Wang, L. Zhao, Y. Liu, et al. , ACPA-negative rheumatoid arthritis: From immune mechanisms to clinical translation. EBioMedicine 83 , 104233 (2022).
[4] D. Sieghart, A. Platzer, P. Studenic, et al. , Determination of autoantibody isotypes increases the sensitivity of serodiagnostics in rheumatoid arthritis. Front. Immunol. 9 (2018).
[5] S. D. Seegobin, M. H. Y. Ma, C. Dahanayake, A, et al. , ACPA-positive and ACPA-negative rheumatoid arthritis differ in their requirements for combination DMARDs and corticosteroids: secondary analysis of a randomized controlled trial. Arthritis Res. Ther. 16 , R13 (2014).
[6] A. C. Boer, A. Boonen, A. H. M. van der Helm van Mil, Is anti-citrullinated protein antibody-positive rheumatoid arthritis still a more severe disease than anti-citrullinated protein antibody-negative rheumatoid arthritis? A longitudinal cohort study in rheumatoid arthritis patients diagnosed from 2000 onwar. Arthritis Care Res. 70 , 987–996 (2018).
[7] L. Padyukov, M. Seielstad, R. T. H. Ong, et al. , Epidemiological Investigation of Rheumatoid Arthritis (EIRA) study group, A genome-wide association study suggests contrasting associations in ACPA-positive versus ACPA-negative rheumatoid arthritis. Ann. Rheum. Dis. 70 , 259–265 (2011).
[8] J. He, Y. Chu, J. Li, Q. Meng, Y. Liu, et al. , Intestinal butyrate-metabolizing species contribute to autoantibody production and bone erosion in rheumatoid arthritis. Sci. Adv. 8 , eabm1511 (2022).
[9] K. Y. Cunningham, B. Hur, V. K. Gupta, et al. , Patients with ACPA-positive and ACPA-negative rheumatoid arthritis show different serological autoantibody repertoires and autoantibody associations with disease activity. Sci. Rep. 13 , 5360 (2023).
Acknowledgements: NIL.
Disclosure of Interests: Jaeyun Sung: None declared, Benjamin Hur: None declared, Vinod Gupta: None declared, Minsik Oh: None declared, Hu Zeng: None declared, Cynthia S. Crowson: None declared, Kenneth J Warrington: None declared, Elena Myasoedova: None declared, Vanessa L. Kronzer: None declared, John M Davis III J.M.D. has a research grant from Pfizer.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (