

Background: Rheumatoid arthritis (RA) typically affects specific joints, with the small joints of the hands (MCP and PIP) being the most involved. In contrast, the shoulder and knee joints are less frequently affected. The mechanisms underlying this characteristic pattern of joint involvement are not elucidated yet. We and others showed that synovial fibroblast (SF) from different joint locations vary in their transcriptome, epigenome, and function [1]. The primary cilium is a microtubule-based structure that emanates from the cell surface of most mammalian cell types, partaking in various signaling processes.
Objectives: Here we aim to decipher the functional and morphological differences in primary cilia of SF from MCP/PIP II-V and knee joints in RA patients.
Methods: Synovial tissues were collected from RA patients who met the 2010 ACR/EULAR criteria for RA classification and underwent joint replacement surgery. Single-cell RNA sequencing (scRNAseq) and bulk RNAseq were performed on freshly isolated RA synovial tissue (MCP/PIP II-V (n=3) and knee (n=6)), and RA cultured SF (MCP/PIP II-V (n=3) and knee (n=5)) respectively. Knee tissues obtained from elective arthroscopy after trauma (n=5) and MCP/PIP tissues obtained from patients following hand surgery (e.g. joint fractures, arthrolysis) (n=6) were used as controls. Primary cilia-associated genes were identified using UniProtKB with the keyword “Cilium” (KW-0969). Hedgehog (Hh) target genes were obtained from CollecTRI [2]. Primary cilia were measured by immunofluorescence staining of acetylated alpha-tubulin in serum-starved RA SFs from knee (n=3) and hand (n=5). A watershed algorithm was used for image segmentation to quantify primary cilia in 3,000 to 5,000 cells per patient. Cultured SFs were stimulated with TNF (10 ng/ml) and Purmorphamine (1 µM). Hh activity was assessed using a GLI reporter dual luciferase assay (control MCP/PIP II-V (n=6), RA MCP/PIP II-V (n=6), control knee (n=5), RA knee (n=7)) and GLI3 processing was analyzed by Western blot (n=3). Chymotrypsin-like, trypsin-like, and caspase-like proteasome activities were measured using the Proteasome-Glo 3-Substrate Cell-Based Assay (MCP/PIP II-V (n=5-7), knee (n=3-4).
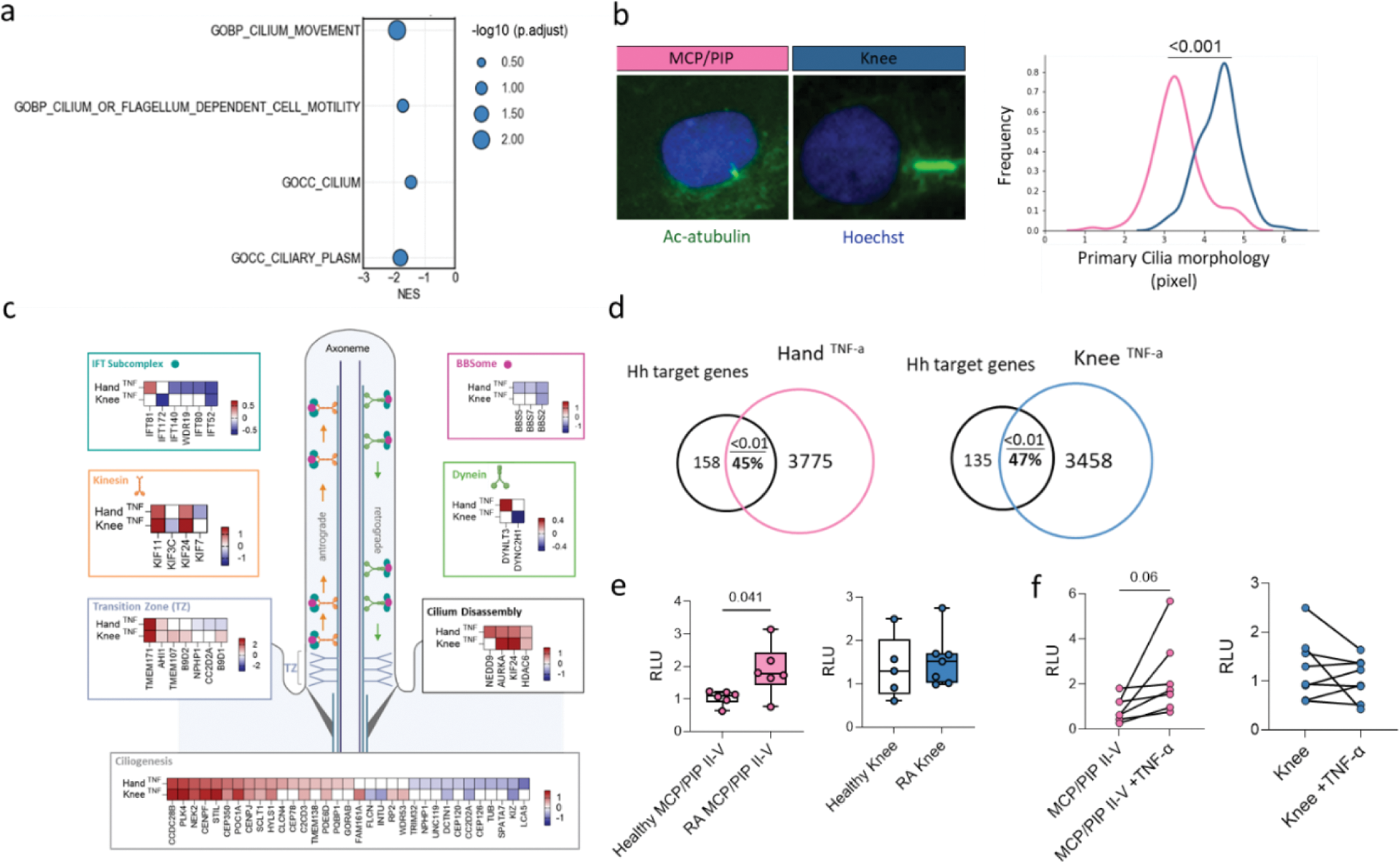
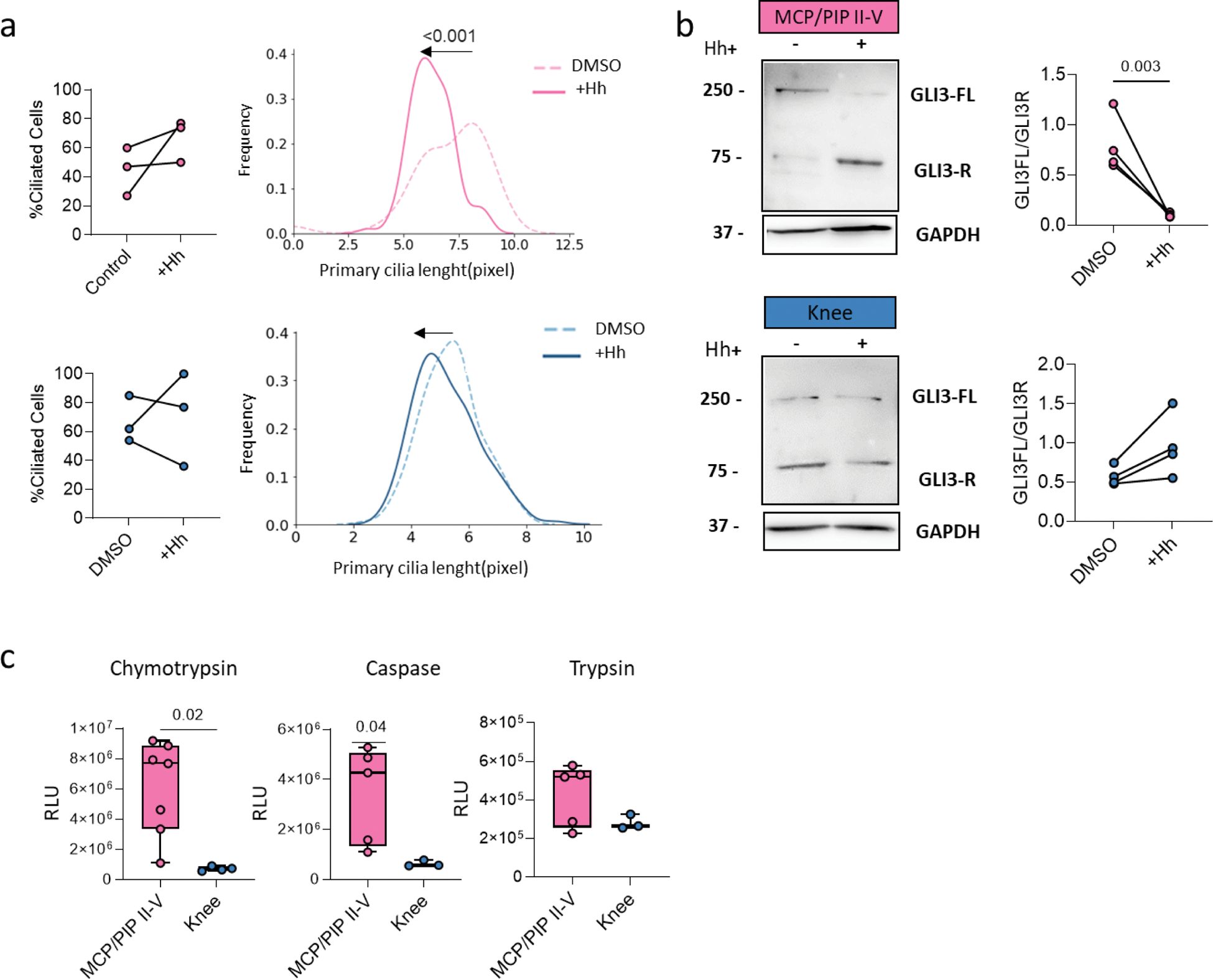
Results: scRNAseq and bulk RNAseq highlighted enrichment of primary cilia-associated terms between MCP/PIP II-V and knee SFs (FDR<0.05, |log2FC| > 20%) (Figure 1a). Cultured SFs in vitro showed longer cilia in knee and shorter in MCP/PIP joints (Figure 1b). Bulk RNAseq of TNF-stimulated SFs indicated differential expression of ciliary architecture (Transition Zone, Ciliogenesis) and trafficking (IFT Subcomplex, BBSome, Kinesin, Dynein) genes, predominantly in MCP/PIP SFs (FDR<0.05, |log2FC|>20%) (Figure 1c). The primary cilium regulates Hh signaling, and RNAseq revealed enriched Hh target genes in unstimulated MCP vs knee SFs (NES=1.5, p.adj=0.01). Despite no activity differences between MCP/PIP II-V and knee SFs, Hh signaling was elevated in MCP II-V RA SFs vs control SFs (p=0.04), but not in knee RA SFs (Figure 1e). TNF stimulation enriched Hh targets in both MCP and knee SFs (p.adj=0.01) (Figure 1d); however, only MCP/PIP SFs showed increased Hh activity in vitro after 24h (p=0.02) (Figure 1f). Purmorphamine-induced Hh signaling did not alter cilia counts but reduced cilia length in MCP/PIP II-V (p<0.001), not knee SFs (Figure 2a). This correlated with downregulated cilia gene signatures post-TNF, linking shorter cilia to higher Hh activity. GLI3 cleavage, a proteasome-mediated process central to Hh signaling, increased in MCP/PIP SFs (p<0.0001) and decreased in knee SFs (p=0.01) post-Hh induction (Figure 2b). Accordingly, proteasome activity, inversely related to cilia length, was higher in MCP/PIP SFs (p<0.05) (Figure 2c).
Conclusion: Our study showed that primary cilia morphology and function, including Hh signaling activity, vary significantly between SFs from different joint locations. Given the importance of primary cilia in various biological processes, these site-specific differences may play a crucial role in regulating fibroblast functions and could influence the development and progression of RA.
REFERENCES: [1] Buckley CD, et al. Location, location, location: how the tissue microenvironment affects inflammation in RA. Nat Rev Rheumatol. 2021;17(4):195-212
[2] Müller-Dott S, et al. Expanding the coverage of regulons for accurate estimation of transcription factor activities. Nucleic Acids Res. 2023;51(20):10934-49.
a ) Overrepresentation analysis of ciliary terms for MCP-Knee DEGs (scRNAseq). b ) Immunostaining of primary cilia (anti-acetylated alpha tubulin, green) in 24hr serum-starved SF. Kolmogorov-Smirnov test. c ) Functional categories of ciliary genes (IFT complex, TZ, ciliogenesis) post-TNF in hand (n=6) and knee (n=4) SFs. Red: upregulated; blue: downregulated (|FC| >20%, FDR < 0.05). d ) Hh target gene enrichment in TNF-stimulated SF (GSEA, adj p-values shown). e ) Hh activity in control/RA MCP/PIP II-V and knee SFs (Mann-Whitney). f ) Hh activity in TNF-stimulated MCP/PIP II-V (n=7) and knee (n=6) SFs at 48hr (Wilcoxon). p-values >0.08 not shown.
a ) Ciliated SF fraction and cilia length distribution after 72hr Purmorphamine (+Hh) in MCP/PIP (pink) and knee (blue). Kolmogorov-Smirnov test. b ) Western blot of GLI3 in +Hh-stimulated MCP/PIP and knee SFs with quantification. c ) Proteasome activity in RA SFs (MCP/PIP II-V, knee). Data: mean ± SD. One-way ANOVA. p-values >0.08 not shown.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (