

Background: Spondyloarthritis (SpA) exhibits strong familial aggregation, with close relatives of patients having a significantly higher risk of developing the disease. This pattern might be driven by an accumulation of common variants that collectively increase disease risk or by rare variants in high linkage disequilibrium (LD) with common risk alleles, amplifying the genetic predisposition in affected families.
Objectives: To investigate whether the familial form of SpA is influenced by an excess burden of common risk variants or by rare variants located within SpA-associated loci.
Methods: We examined two independent patient groups: (1) a family-based cohort of 376 SpA patients from 68 French families with at least four affected members, all of whom underwent whole genome sequencing (WGS), and (2) a cohort of 242 French unrelated ankylosing spondylitis (AS) patients with and without a family history of SpA, genotyped on the Illumina CoreExome array. Polygenic risk score (PRS) for AS were calculated using a model developed by Li et al. for Western European population [1]. Linear regression was applied to identify PRS-associated factors, and HLA-B27 frequency differences between sporadic and familial cases were analyzed using the chi-square test. We identified 46 coding genes overlapping with SNPs in high LD (r²>0.8) with lead SNPs from the 42 known AS-associated loci [2]. To examine whether rare variants within these genes contribute to familial SpA, we conducted a rare variant burden test in our family cohort using gnomAD data as controls [3].
Results: PRS was significantly increased in patients with familial history of SpA (68 probands from multiplex families and 123 patients from the unrelated cohort) compared to 119 sporadic cases (β-estimate (β)± standard error (SE): 0.09 ± 0.03; p-value (p) = 0.004; Figure 1A). However, the HLA-B27 positivity rate was significantly higher in familial cases compared to sporadic cases (94.2% vs 78.2%; p = 6x10 -5 ) and the PRS difference was no longer significant when comparing only HLA-B27 positive patients (β ± SE: 0.003 ± 0.03; p = 0.9; Figure 1B). Within the familial cohort, PRS was inversely associated with the number of affected family-members (β ± SE: -0.08 ± 0.03; p = 0.03). WGS data from the familial cohort revealed at least one non-synonymous SNPs with a minor allele frequency below 1% in non-Finnish European population and a predicted pathogenicity CADD score above 20 in 19 of the 46 AS-associated coding genes. The rare variant-burden test showed nominal association with familial SpA only for the PDE4A gene (p=0.05).
Distribution of PRS according to family history status in all SpA patients (panel A) or in HLA-B27 positive patients only (panel B)
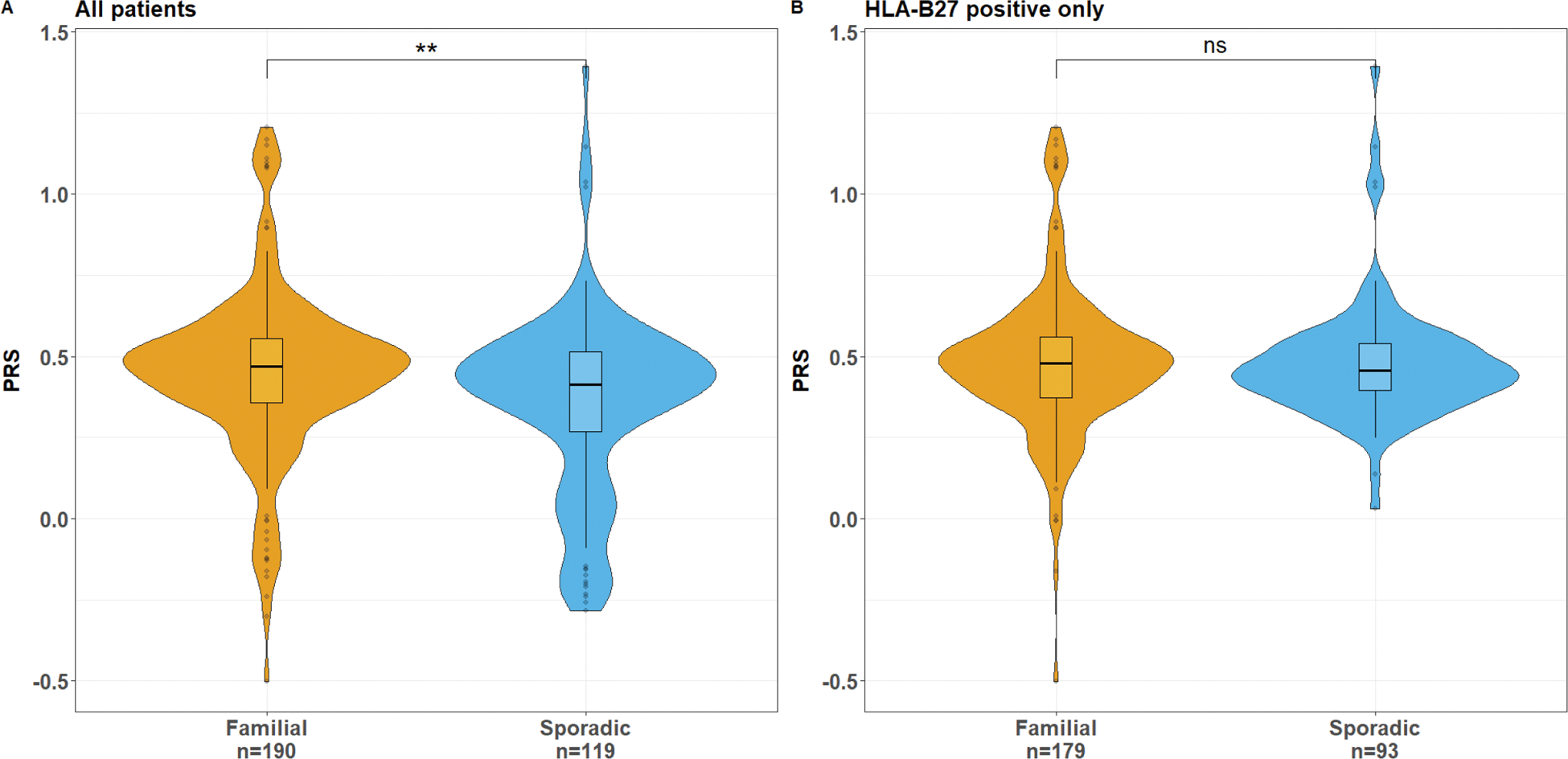
Conclusion: Besides the higher prevalence of HLA-B27, we found no significant accumulation of common AS-associated variants in familial cases of SpA. Furthermore, there was no excess of rare variants in genes within AS-associated loci, with the exception of a nominal association for PDE4A . Altogether these results suggest the involvement of other genetic factors, such as rare variants in additional genes, or environmental factors contributing to the strong familial aggregation of SpA.
REFERENCES: [1] Li Z, Wu X, Leo PJ, De Guzman E, Akkoc N, Breban M, et al. Polygenic Risk Scores have high diagnostic capacity in ankylosing spondylitis. Ann Rheum Dis 2021;80:1168–74.
[2] Ellinghaus D, Jostins L, Spain SL, Cortes A, Bethune J, Han B, et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet 2016;48:510–8.
[3] Lali R, Chong M, Omidi A, Mohammadi-Shemirani P, Le A, Cui E, et al. Calibrated rare variant genetic risk scores for complex disease prediction using large exome sequence repositories. Nat Commun 2021;12:5852.
Acknowledgements: This project was supported by an unrestricted grant from UCB Pharma.
Disclosure of Interests: Félicie Costantino Amgen, Jansenn, Lilly, Celltrion, Abbvie, Novartis, UCB, UCB, Roula SAID: None declared, Hendrick Mambu-Mambueni: None declared, Anne Boland: None declared, Jean-François Deleuze: None declared, Matthew Brown Clementia, Grey Wolf, Incyte, Ipsen, Altis Medicines, UCB, Henri-Jean Garchon: None declared, Maxime Breban UCB, Novartis, UCB, MSD, Novartis.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (