

Background: Ankylosing spondylitis (AS) is a chronic inflammatory disease that primarily targets the axial skeleton, particularly the spine and sacroiliac joints, often resulting in pain, stiffness, and potential fusion of vertebrae [1]. AS pathogenesis is increasingly linked to immune dysregulation, yet the precise cellular mechanisms remain unclear. Immune cells, particularly T cells, appear to play a significant role in chronic inflammation and tissue remodeling observed in AS, as do distinct immune cell profiles and clonally expanded T cells within affected tissues [2, 3]. This study aims to comprehensively characterize the immune landscape in AS by exploring immune cell subsets in both peripheral blood and affected tissues, such as synovium fluid and spine ligaments, to uncover potential mechanisms underlying disease progression.
Objectives: The primary goal of this study was to investigate the immunological characteristics of AS, with a focus on T cell subpopulations in peripheral blood and affected joint tissues. Specifically, we aimed to:
Compare immune profiles between AS patients and healthy controls (HDs).
Identify immune cell subtypes that contribute to chronic inflammation in AS.
Examine clonal expansion and T cell receptor (TCR) gene usage across synovium fluids and spine ligaments.
Methods: The study involved 40 AS patients, classified by disease activity, and 100 HDs. Peripheral blood samples were analyzed using multicolor flow cytometry, with 51 antibodies targeting cell lineage and functional markers to assess immune cell subpopulations. T-distributed stochastic neighbor embedding (TSNE) analysis was applied to visualize immune profiles. To further study immune responses in affected tissues, synovium fluid and spine ligament samples were collected from AS patients. Single-cell RNA sequencing (scRNA-seq) and T cell receptor sequencing (TCR-seq) were performed on CD3+ T cells using the 10X Genomics Chromium platform, supplemented by public datasets of AS synovium fluids and peripheral immune cells from HDs. Unsupervised uniform manifold approximation and projection (UMAP) analysis classified T cells into major subpopulations, and gene set enrichment analysis (GSEA) was used to identify activated pathways.
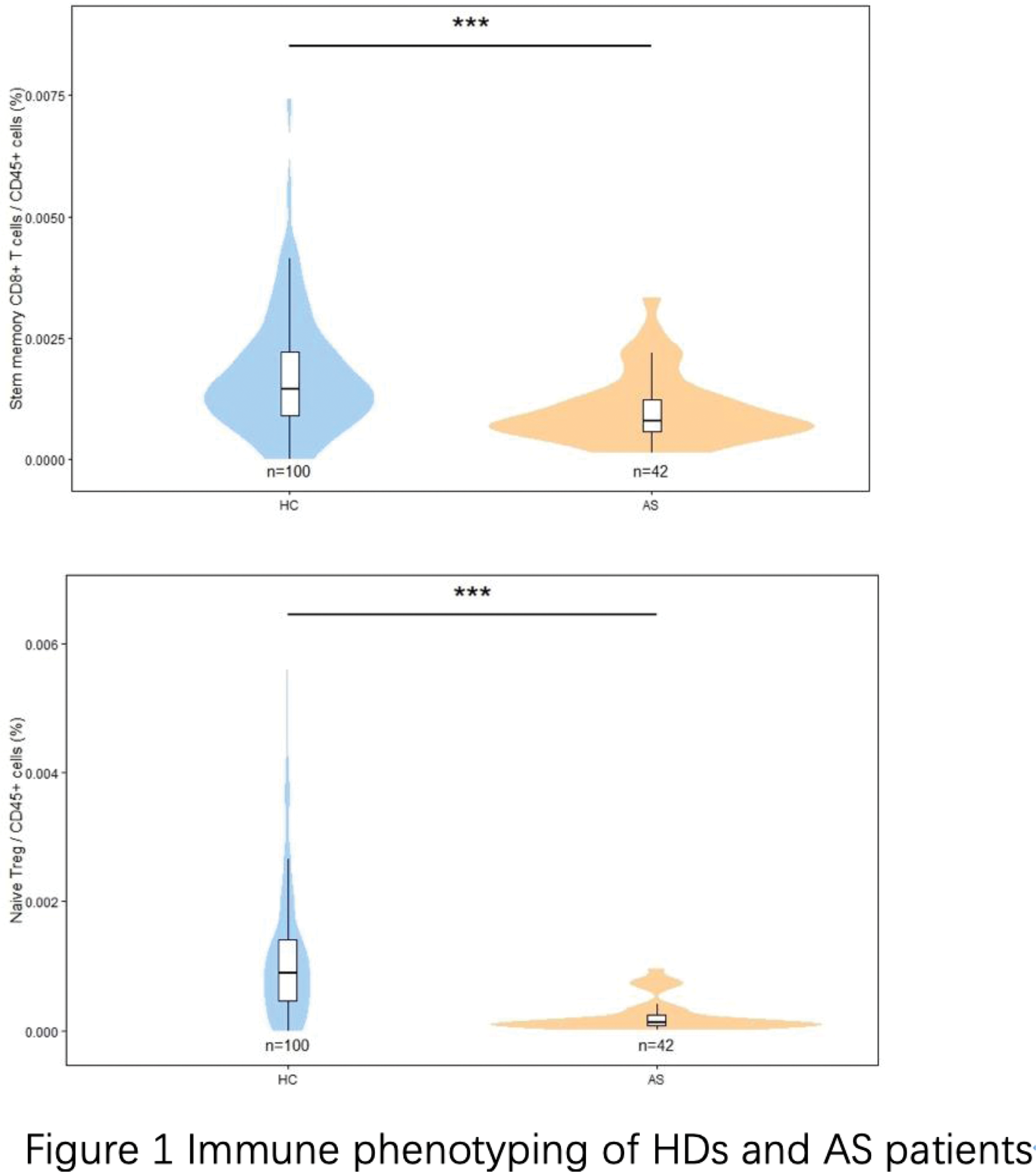
Peripheral Blood Analysis : TSNE analysis revealed distinct immune cell profiles in AS patients versus HDs, especially in CD8+ T cells and Treg cells. Early T cell types, such as stem memory T cells and naive Tregs, were decreased in AS patients compared to controls. These naive T cells also showed an inverse correlation with AS Disease Activity Score with C-reactive Protein (ASDAS-CRP), suggesting their depletion in chronic inflammation. This depletion was mirrored in age-related declines of naive Tregs and stem memory T cells, indicating that chronic inflammation in AS may lead to continuous activation and aging of immune resources (Figure 1).
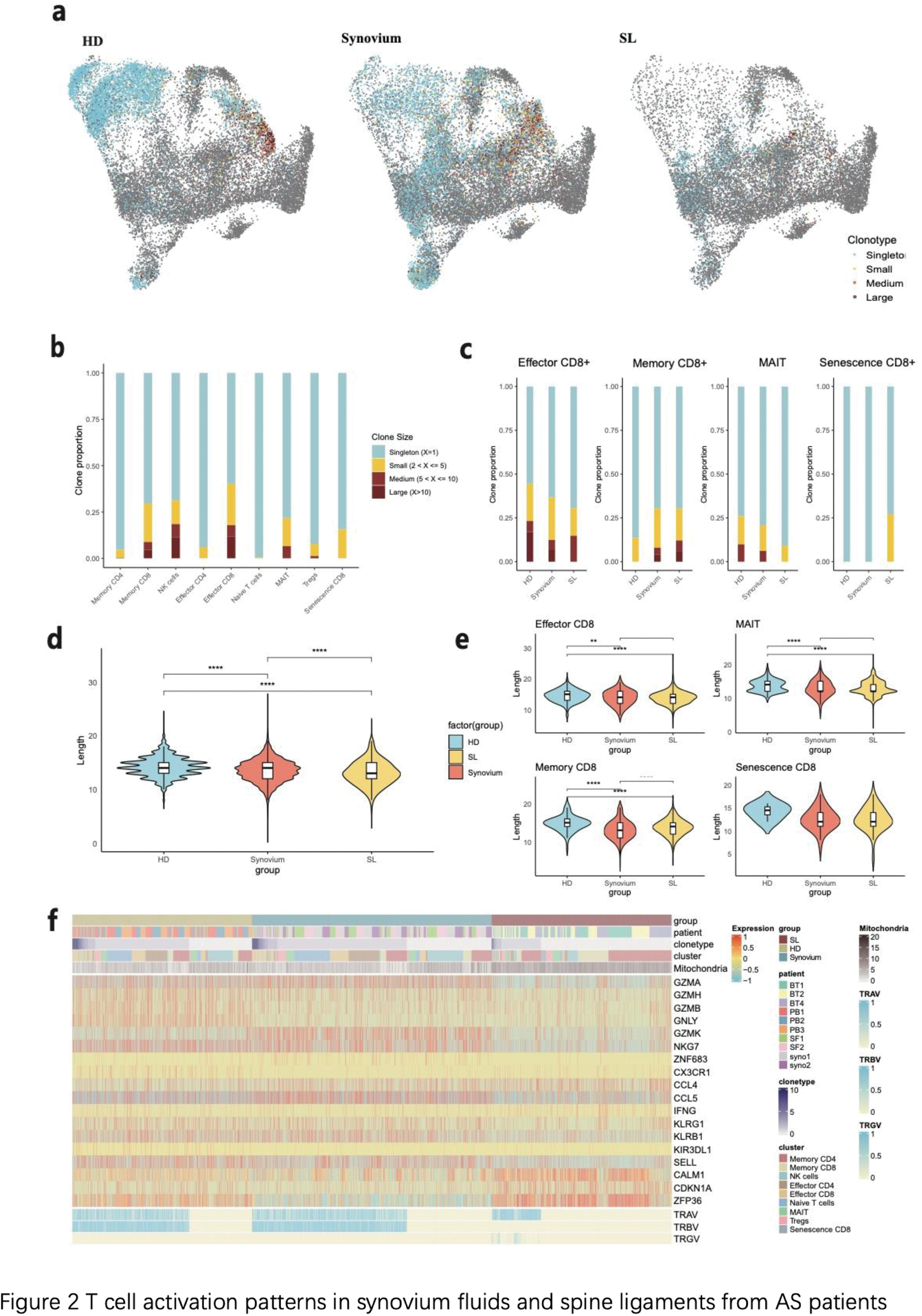
Synovium Fluid and Spine Ligament Analysis : Single-cell sequencing identified nine major T cell clusters, including naive, memory, and effector CD4+ and CD8+ T cells, as well as MAIT cells, NK cells, and Tregs. Notably, memory cells were enriched in AS-affected axial joints, while MAIT cells and Tregs were found exclusively in synovium fluids. DGE analysis showed distinct transcriptional profiles across these cell types, with GSEA identifying unique activation of the T cell receptor signaling pathway, TH1 and TH2 differentiation in MAIT cells from synovium fluids, and T cell differentiation in memory CD8+ T cells in synovium fluids and spine ligaments (Figure 2).
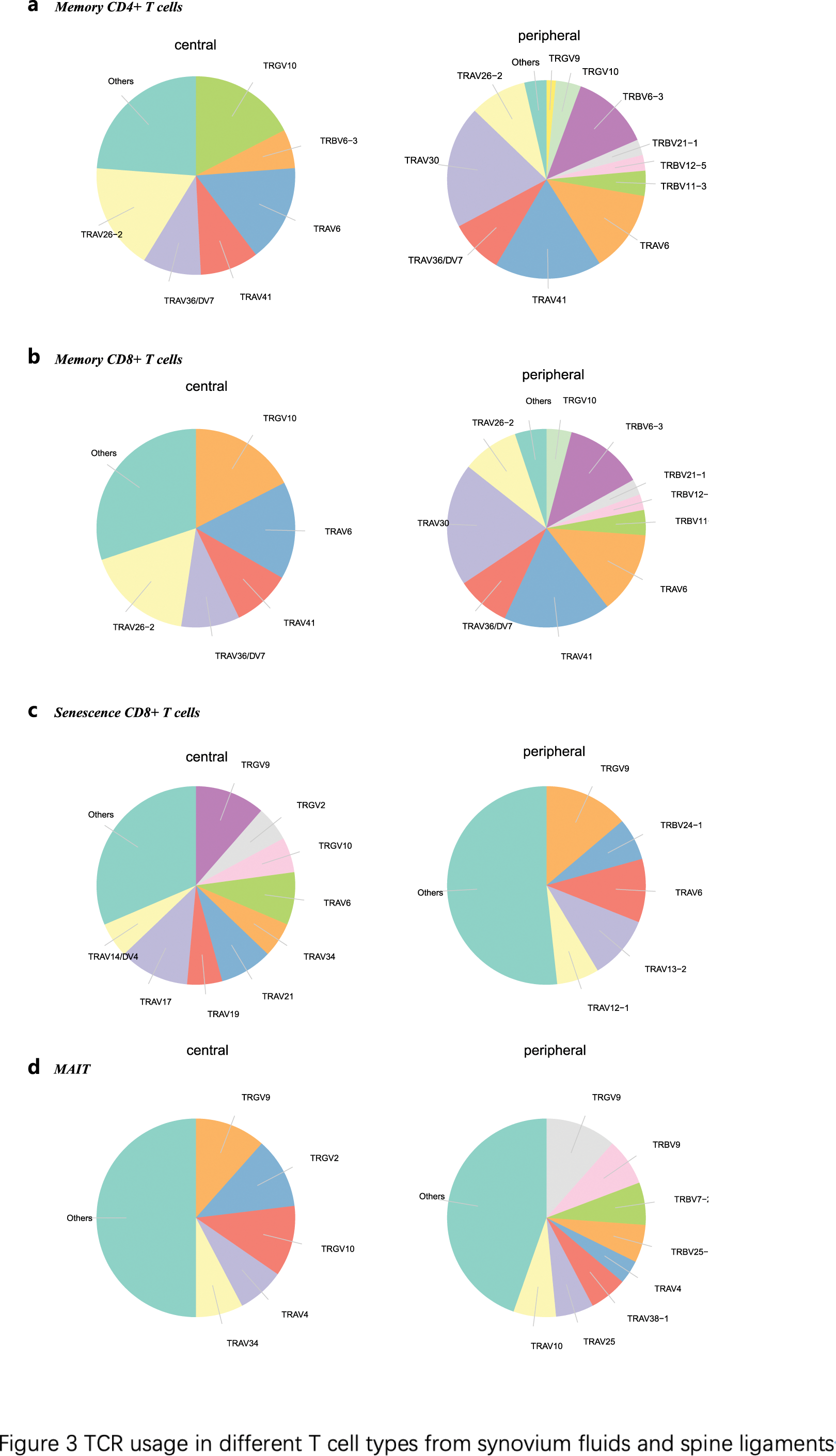
Clonal Expansion and TCR Usage : Single-cell TCR sequencing revealed higher clonal expansion in synovium fluids compared to spine ligaments. In particular, CD8+ T memory and effector cells showed significant clonal expansion in synovium fluids, along with MAIT and Treg cells, indicating an active, tissue-specific immune response. TCR gene analysis revealed differential usage: TRGV genes, specifically TRGV9 and TRGV10, were highly utilized in spine ligaments, while synovium fluids exhibited diverse TRAV usage, with TRAV30 and TRAV41 among the most prominent (Figure 3).
Conclusion: This study presents a comprehensive immune profiling of AS, revealing distinct immune cell dynamics in peripheral blood and affected joint tissues. AS is characterized by systemic immune dysregulation, reflected in the depletion of naive immune cells and increased clonal expansion of memory and effector T cells in the synovium fluids. Furthermore, the differential TCR gene usage between synovium fluids and spine ligaments underscores the possibility of tissue-specific antigenic responses in AS pathogenesis. These findings advance our understanding of immune mechanisms in AS and suggest that targeted modulation of T cell responses, particularly in synovium fluids, may hold therapeutic potential. Future research should investigate the antigenic drivers of clonal expansion and explore targeted immune interventions to mitigate disease progression in AS.
REFERENCES: [1] Ranganathan V, Gracey E, Brown MA, Inman RD, Haroon N. Pathogenesis of ankylosing spondylitis - recent advances and future directions. Nat Rev Rheumatol . Jun 2017;13(6):359-367. doi:10.1038/nrrheum.2017.56
[2] Yang X, Garner LI, Zvyagin IV, et al. Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides. Nature . Dec 2022;612(7941):771-777. doi:10.1038/s41586-022-05501-7
[3] Yi K, Jo S, Song W, et al. Analysis of Single-Cell Transcriptome and Surface Protein Expression in Ankylosing Spondylitis Identifies OX40-Positive and Glucocorticoid-Induced Tumor Necrosis Factor Receptor-Positive Pathogenic Th17 Cells. Arthritis Rheumatol . Jul 2023;75(7):1176-1186. doi:10.1002/art.42476
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (