

Background: We found a population of CD4 + HLADR + T cells in blood that were shown previously to immunologically mirror that of the inflammatory and TCR profile of synovium T cells [1, 2]. These circulatory T cells were shown to be elevated in active disease arthritic individuals and in patients non-responsive to TNF inhibitor treatment. How CD4 + HLADR + effector and regulatory T cells contribute overall to arthritic synovial pathogenicity and their disease relations, remain unknown.
Objectives: This study (Figure 1) seek to elucidate the functional and pathogenic properties of CD4 + HLADR + T cells in juvenile idiopathic arthritic (JIA) patients, through (a) high parametric interrogation of circulatory and synovial immune subsets with the aid of mass cytometry (CyToF), (b) transcriptomic examination of pathogenic pathways with deep RNA sequencing and (c) regulatory and antigenic T cell assays to dissect disease mechanisms.
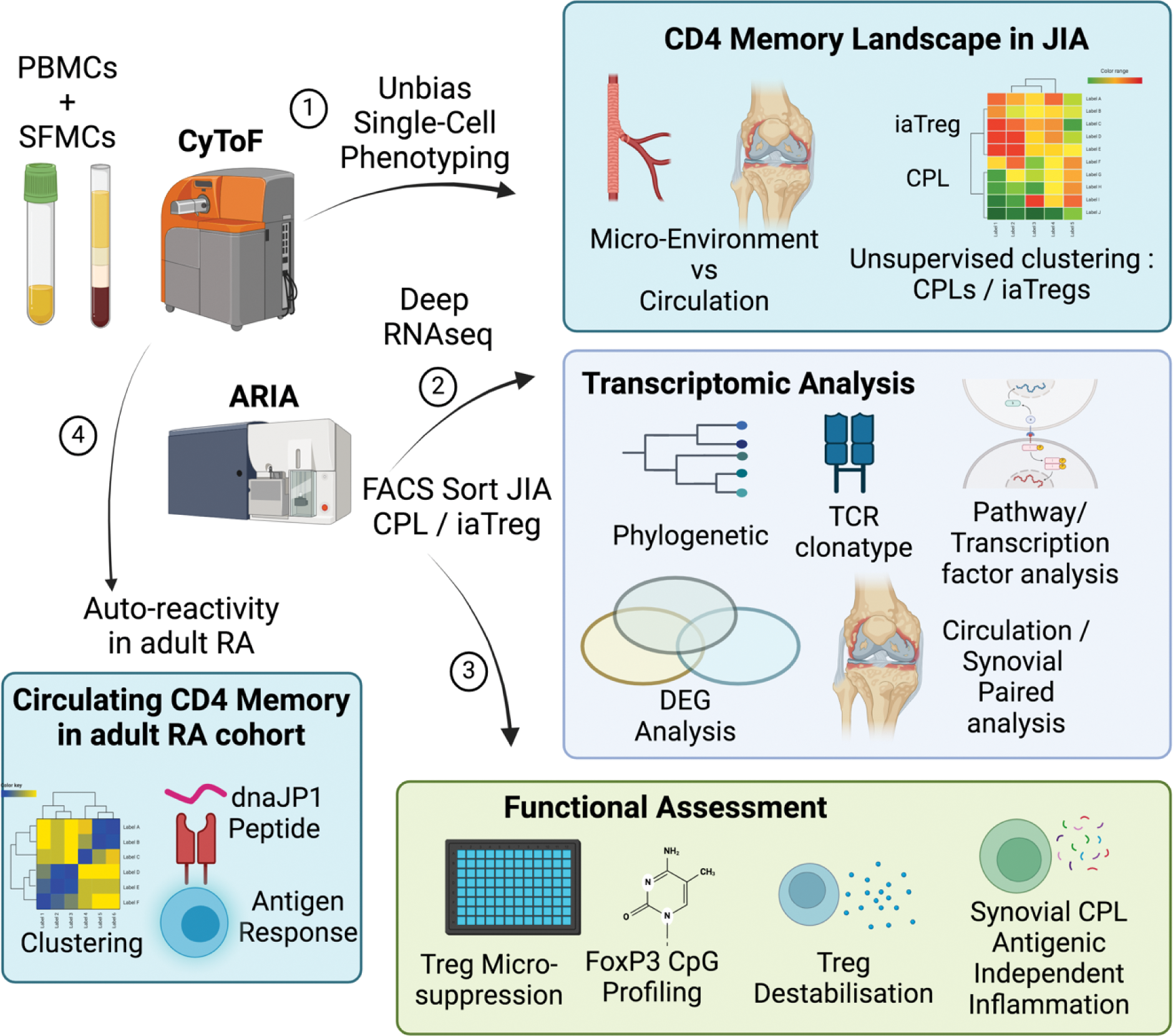
Methods: Mass cytometry interrogation of JIA patients (7 PBMCs, 7 SFMCs), RA patients (10 PBMCs) and healthy controls (10 PBMCs) and analysed with EPIC analytics suite [3]. FACS sorted for CD4 + HLADR ± effectors or regulatory T cells from JIA patients (7 PBMCs, and n=8 paired PBMCs/SFMCs). CpG methylation profiling of FoxP3 promoter site in Tregs and T effectors from JIA patients (3 paired PBMCs/SFMCs) and healthy controls (6 PBMCs). Flow cytometry analysis of JIA (5 PBMCs) and healthy controls (11 PBMCs). Functional assays; (a) Treg suppression assay in JIA patients (8 paired PBMCs/SFMCs) and healthy controls (7 PBMCs), (b) Treg destabilisation assay (n=6), (c) antigenic memory recall response of JIA (7 SFMCs).
Results: We performed an unbiased interrogation of the CD4 + CD45RO + memory landscape of JIA patients with mass cytometry, and detected a up-regulation of CD4 + HLADR + T effectors (p <0.001) within the synovium as compared to blood. Importantly, the synovium CD4 + HLADR + T effectors were highly inflamed as compared with HLADR- counterparts, expressing higher levels of inflammatory cytokines (TNFa, IFNg and IL-21). Notably, CD4 + HLADR + T effectors and regulatory T cells positively correlated (r=0.74, p < 0.01) across spatial disease sites (blood versus synovium compartments), suggesting a micro-environmental relation, whereas CD4 + HLADR - subsets did not. Next, to examine and dissect the functional properties and relationships between CD4 + HLADR + subsets, we sorted JIA PBMCs/SFMCs and performed deep RNA sequencing. Intriguingly, phylogenetic tree analysis reveal a closer relation between circulatory CD4 + HLADR + T effectors and regulatory cells as compared with their original CD4 + effector or regulatory compartments, indicating strong disease convergence. This phenomenon of transcriptomic convergence between CD4 + HLADR + effector and regulatory subsets was further substantiated by pathway and transcription factor network analysis, reflecting similar mega T cell activation pathways and transcription factor drivers. Closer inspection of CD4 + HLADR + subsets within the synovium revealed an enrichment for 7 key pathways (MHC-II, IFNg signalling, viral defence, chemotaxis, apoptosis, T cell co-stimulation and LPS/Bacteria defence). Prominently, HLADR + effector and regulatory subsets in synovium demonstrated strong TCR clonatype sharing (p < 0.01) as compared with HLADR - counterparts. While synovium CD4 + HLADR + Tregs indicated a stable CpG methylation lineage profile in the FoxP3 TSDR promoter site, and were poised to suppress disease T effectors, CD4 + HLADR + T effectors were able to resist their regulatory control (p < 0.05). This is further underscored by the ability of polarising Th-1 environment to destabilise FoxP3 levels (p < 0.05) in CD4 + HLADR + Tregs. Furthermore, JIA synovium CD4 + HLADR + T effectors were able to elicit inflammatory autoimmune memory recall response towards disease relevant peptide HSP60 in an independent manner. Additionally, autoimmune reactivity of CD4 + HLADR + T cells towards the dnaJP1 peptide was demonstrated in adult rheumatic disease individuals, indicating their disease relevance in adult populations.
Conclusion: CD4 + HLADR + T effectors were expanded and highly inflammatory within the synovium compartment. Notably, CD4 + HLADR + Tregs demonstrated strong transcriptomic convergence and TCR clonatype sharing with their analogous HLADR + effectors, indicating a common disease origin. While CD4 + HLADR + Tregs were lineage stable and capable of suppression, CD4 + HLADR + effectors could circumvent regulatory control and elicit an independent inflammatory response to disease relevant antigens. CD4 + HLADR + T effectors may present themselves as an amenable therapeutic target in autoimmune arthritis.
REFERENCES: [1] Spreafico R and et al. A circulating reservoir of pathogenic-like CD4+ T cells shares a genetic and phenotypic signature with the inflamed synovial micro-environment. Ann Rheum Dis. 2016 Feb;75(2):459-65. DOI: 10.1136/annrheumdis-2014-206226.
[2] Rossetti M, and et al. TCR repertoire sequencing identifies synovial Treg cell clonotypes in the bloodstream during active inflammation in human arthritis. Ann Rheum Dis. Feb 2017; 76(2): 435-441. DOI: 10.1136/annrheumdis-2015-208992.
[3] Yeo JG, and et al. The Extended Polydimensional Immunome Characterization (EPIC) web-based reference and discovery tool for cytometry data. Nat Biotechnol. 2020 Jun;38(6):679-684. DOI: 10.1038/s41587-020-0532-1. Erratum in: Nat Biotechnol. 2020 Jun;38(6):757. DOI: 10.1038/s41587-020-0574-4. PMID: 32440006.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (