

Background: Lupus nephritis (LN) is characterized by complex immune infiltration patterns within the renal microenvironment, with a high heterogeneity among patients.
Objectives: This study aims to spatially characterize immune infiltrates in LN, shedding lights into differences between histological classes and kidney areas such as glomeruli and tubules.
Methods: Immune cell distributions in 22 regions of interest (ROIs) from eight LN kidney biopsies (three class III, five class IV) were assessed using imaging mass cytometry. Furthermore, spatial transcriptomics analysis on two patients was performed to define key molecular pathways at play in LN.
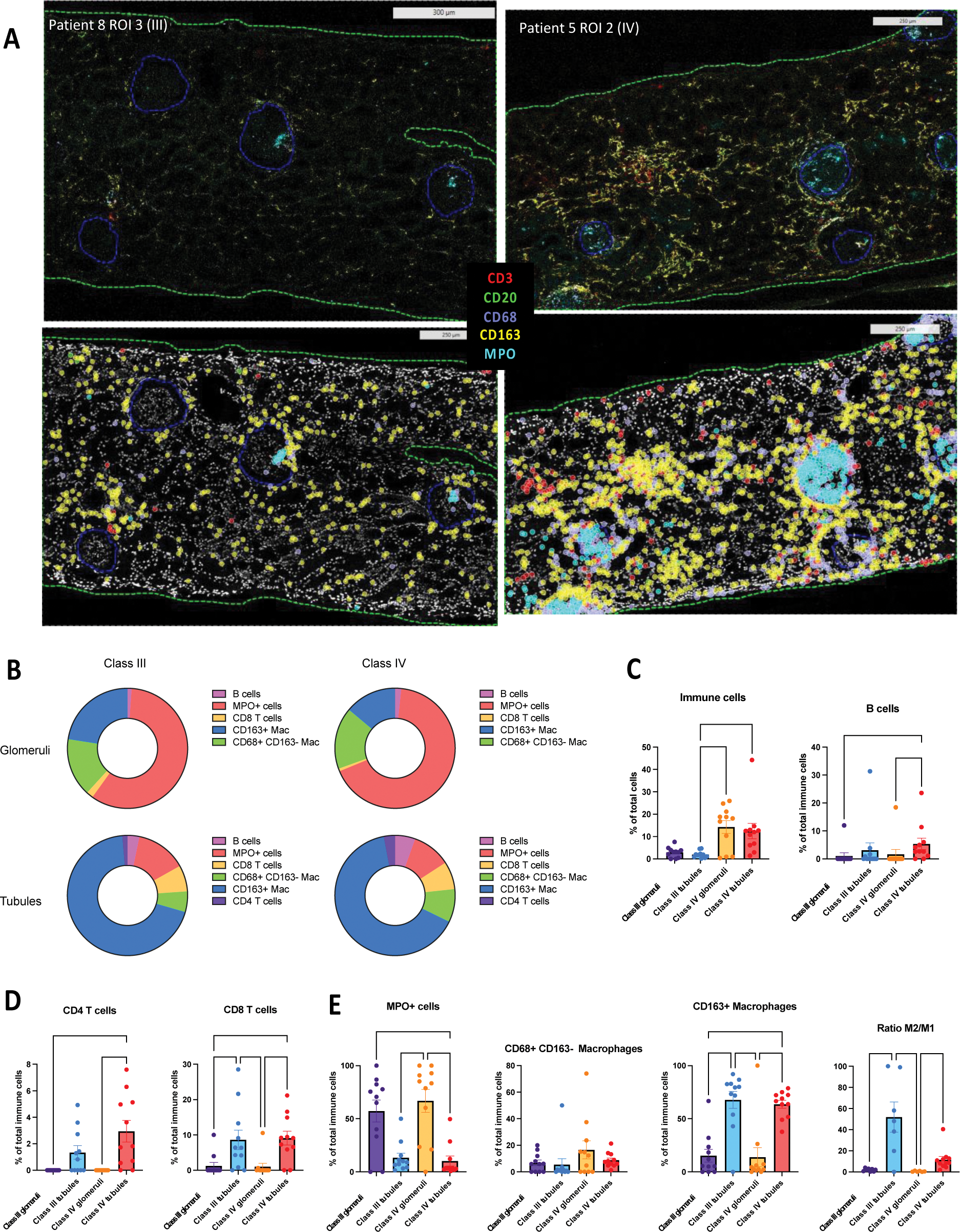
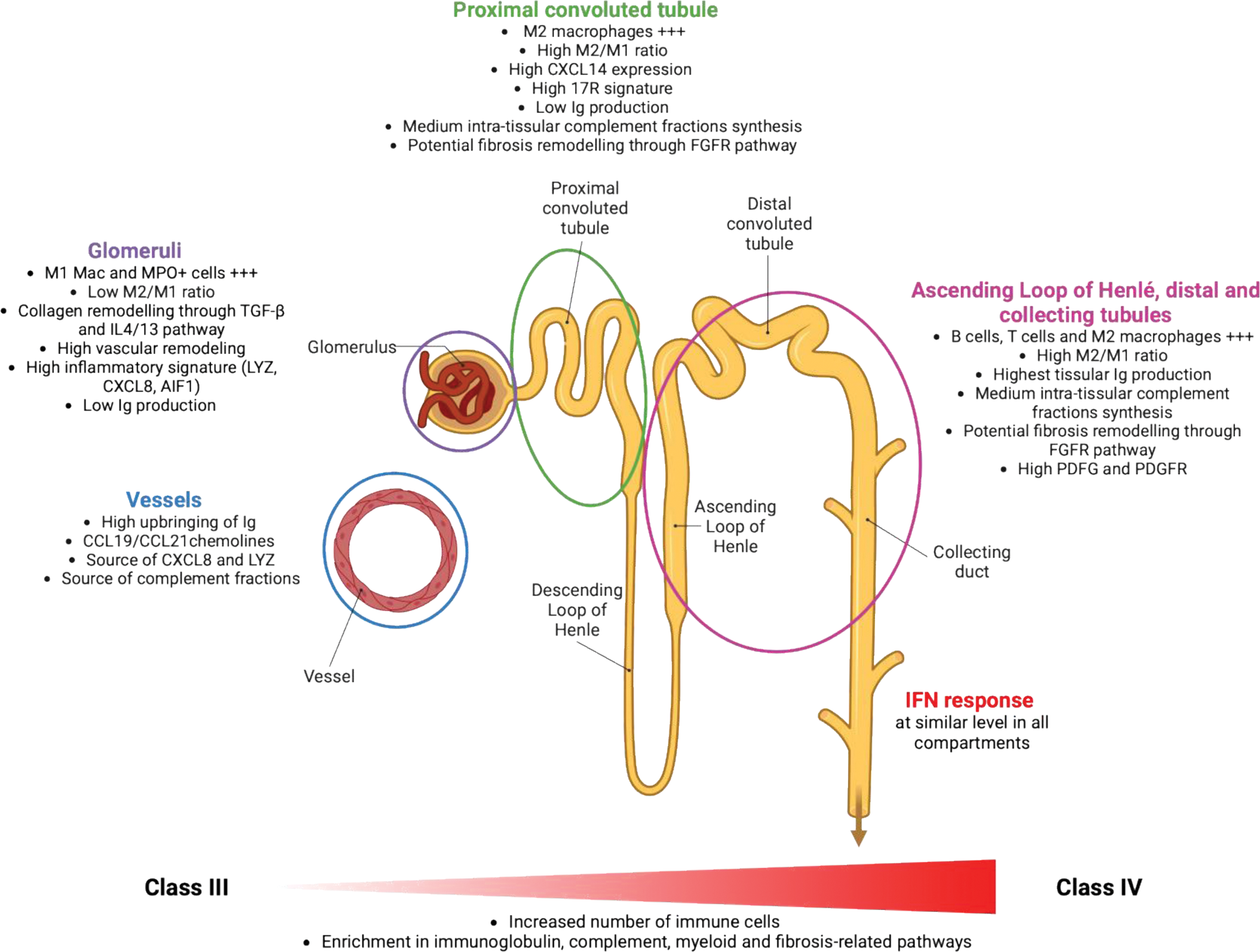
Results: Supervised analysis revealed significantly greater immune cell infiltration in class IV compared to class III ROIs, with distinct immune cell distribution patterns across kidney compartments. Indeed, PMN and CD68+ macrophages (“M1-like”) were enriched in glomeruli, while T cells, B cells and CD163+ macrophages (“M2-like”) predominated in tubules, resulting in a higher M2/M1 ratio in tubules compared to glomeruli (Figure 1). Unsupervised analysis further revealed distinct immune niches, located within distinct renal compartments. Notably, dense and complex immune aggregates of B cells, T cells and myeloid cells were predominantly found in the distal tubules, whereas macrophages were found diffusely throughout all tissue areas, exhibiting various phenotypes according to each kidney location. Spatial transcriptomics finally revealed key-compartment specific pathways: (i) glomeruli exhibited a complex signature with both inflammatory and pro-fibrotic pathways such as IL4/IL13 and TGF-β pathways, (ii) proximal tubules displayed a strong and specific IL17 signature, and (iii) Loop of Henlé, distal and collecting tubules showed intra-tissular immunoglobulin production and fibrotic remodeling via FGFR pathway activation.
Conclusion: This spatially resolved analysis provides novel insights into the heterogeneity of the immune landscape in LN, uncovering distinct class- and compartment-specific immune cell distribution and molecular pathways (Figure 2). These findings offer valuable perspectives to guide future targeted therapeutic strategies.
Funding: This work was supported by an unrestricted grant from GlaxoSmithKline (GSK, study ID 217408).
REFERENCES: [1] Anders HJ, Saxena R, Zhao M hui, Parodis I, Salmon JE, Mohan C. Lupus nephritis. Nat Rev Dis Primers. 23 janv 2020;6(1):1‑25.
[2] Weening JJ, D’Agati VD, Schwartz MM, Seshan SV, Alpers CE, Appel GB, et al. The classification of glomerulonephritis in systemic lupus erythematosus revisited. J Am Soc Nephrol. févr 2004;15(2):241‑50.
[3] Arazi A, Rao DA, Berthier CC, Davidson A, Liu Y, Hoover PJ, et al. The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol. juill 2019;20(7):902‑14.
[4] Der E, Suryawanshi H, Morozov P, Kustagi M, Goilav B, Ranabothu S, et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat Immunol. juill 2019;20(7):915‑27.
[5] Tang PMK, Nikolic-Paterson DJ, Lan HY. Macrophages: versatile players in renal inflammation and fibrosis. Nat Rev Nephrol. mars 2019;15(3):144‑58.
[6] Danaher P, Hasle N, Nguyen ED, Roberts JE, Rosenwasser N, Rickert C, et al. Childhood-onset lupus nephritis is characterized by complex interactions between kidney stroma and infiltrating immune cells. Science Translational Medicine. 27 nov 2024;16(775):eadl1666.
[7] Wu C, Jiang S, Chen Z, Li T, Gu X, Dai M, et al. Single-cell transcriptomics reveal potent extrafollicular B cell response linked with granzyme K+ CD8 T cell activation in lupus kidney. Ann Rheum Dis. 17 oct 2024;ard-2024-225876.
[8] Chen W, Jin B, Cheng C, Peng H, Zhang X, Tan W, et al. Single-cell profiling reveals kidney CD163+ dendritic cell participation in human lupus nephritis. Annals of the Rheumatic Diseases. 1 mai 2024;83(5):608‑23.
Comparison of immune infiltrates according to histological class and kidney area
A. Representative ROIs displaying tissular segmentation for glomeruli and tubulo-interstitial compartment. On the upper side, real ROIs are shown for patients. On the lower side, gating strategy is applied to display the main immune cells (T cells are shown by CD3 in red; B cells are shown by CD20 in green; M1 macrophages are shown by CD68 in lavender; M2 macrophages are shown by CD163 in yellow, and PMN and MPO+ Mac are shown by MPO in turquoise). On the left is one ROI from a class III patient, and on the right is displayed one ROI from a class IV patient. B. Circle plot showing repartition of immune cells according to LN class and renal area. C. Proportion of total immune cells and B cells according to LN class and renal area. D. Proportion of CD4+ T cells and CD8+ T cells according to LN class and renal area. E. Proportion of myeloid cells and M2/M1 ratio according to LN class and renal area.
Graphical abstract
Informed Consent: Informed consent was obtained from all patients included in the study.
Acknowledgements: We would like to thank all the staff of the LBAI and the pathology department of CHU of Brest for their help. We also thank all the members of the Hyperion platform (LBAI, Brest, France) for their assistance.
Disclosure of Interests: Eleonore Bettacchioli This work was supported by an unrestricted grant from GlaxoSmithKline (GSK, study ID 217408), Christelle Le Dantec: None declared, Amandine Meudec: None declared, Amelie Bourhis: None declared, Patrice Hémon: None declared, Marion Le Rochais: None declared, Akhiya A R: None declared, Marie Frutoso: None declared, Arnaud Uguen: None declared, Catherine Hanrotel Saliou: None declared, Sandrine Jousse-Joulin: None declared, Noemie JOURDE-CHICHE: None declared, Divi Cornec: None declared, Sophie Hillion: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (