

Background: Immunophenotypic and transcriptomic studies have revealed shared immune dysregulation in ANA-positive at-risk individuals, whether they develop RMDs or maintain benign autoimmunity. However, functional protein studies are limited, and predictive biomarkers for RMD progression remain unidentified. Biomarkers to identify high-risk individuals for closer monitoring, as well as appropriate targeted therapeutic interventions in subclinical autoimmunity, could offer a novel strategy for SLE prevention.
Objectives: To investigate functional protein dysregulation in ANA-at-risk individuals and identify biomarkers and biological pathways predictive of disease progression.
Methods: The At-Risk study recruited ANA-positive patients with non-specific symptoms who did not fulfil the criteria for ANA-RMDs. Patients were followed for 3 years for a diagnosis of SLE, SjD or other ANA-RMD. We included baseline samples from 103 ANA-at-risk individuals (32 progressors and 67 non-progressors), a follow-up sample from progressors, healthy and established SLE controls. Samples were analysed using Proximity Extension Assay (PEA) proteomics with EXPLORE Inflammation I and II panels (Olink®, Uppsala, Sweden). We selected 193 differently expressed proteins (DEPs) of 620 with ≥0.5 fold difference and adjusted p<0.05 between ANA at risk and HC (n = 10) for further comparison and Weighted Gene Co-expression Network Analysis (WGCNA). Pearson test was used to examine the correlation of WGCNA modules with clinical traits of interest as well as a gene (protein) significance (GS) value for each protein in the module. Protein enrichment analysis was performed to determine the module’s biological significance. Proteins from modules associated with progression were evaluated singly and in combination using AUC. Cytoscape (version 3.10.1) and R version 4.3.1 were employed for data analysis and visualisation.
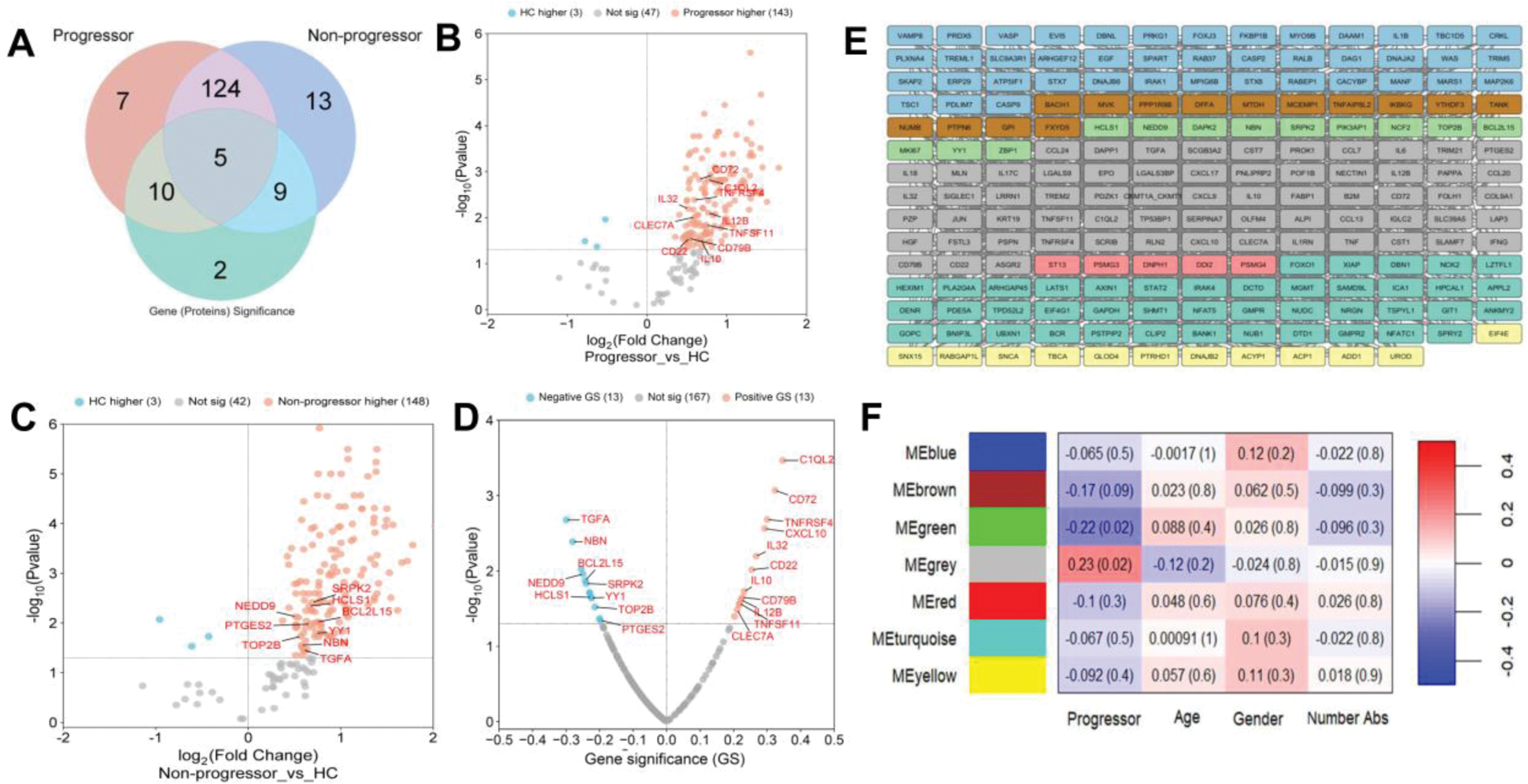
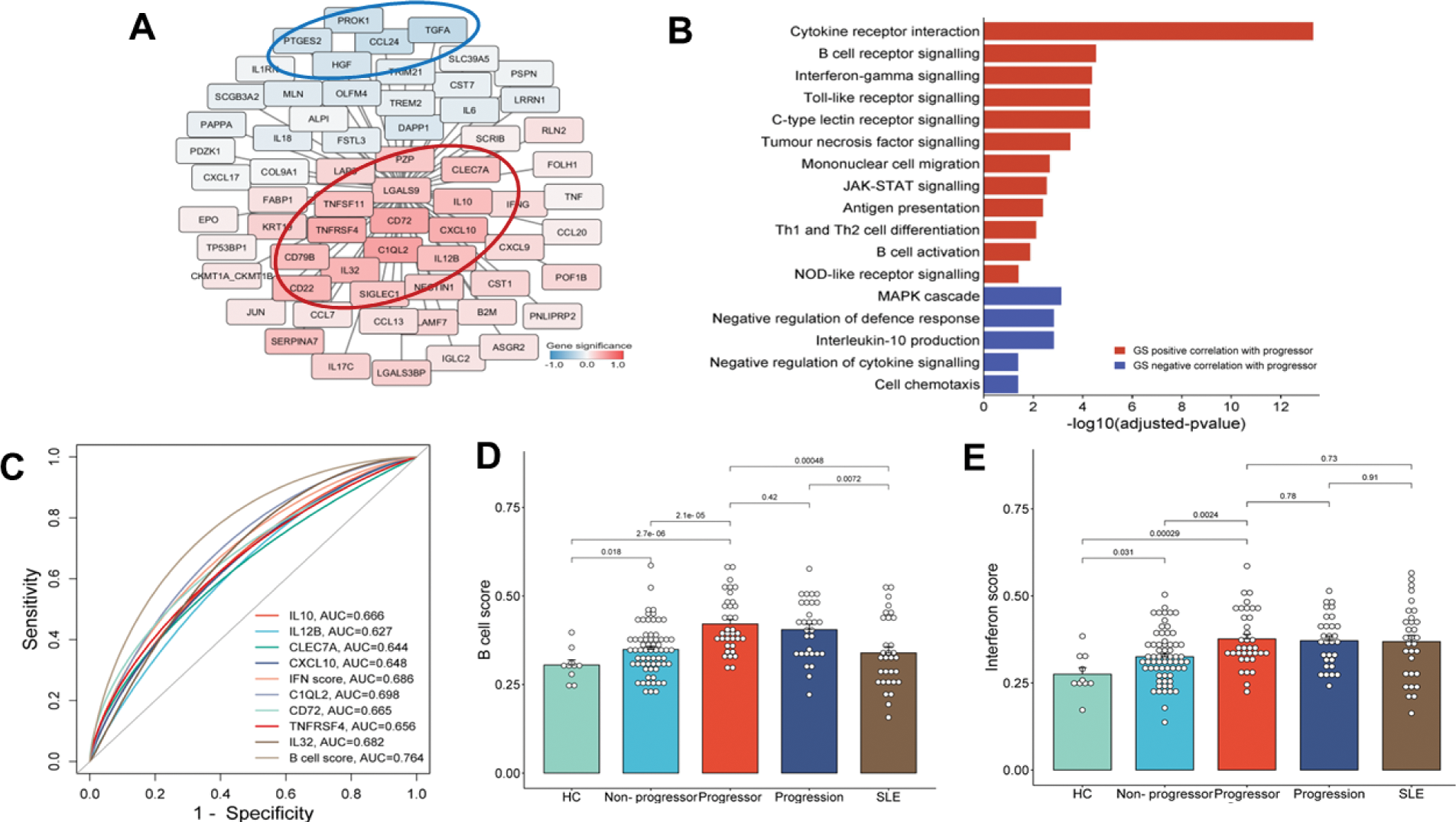
Results: Comparing At-Risk and HC, 168 significant DEPs were identified; 146 in progressors and 151 in non-progressors (Figure 1B, 1C). Of these, 17 and 22 were unique to progressors and non-progressors, respectively (Figuer 1A). WGCNA revealed seven co-expressed protein modules (Figure 1E), with 26 proteins exhibiting adjusted GS p<0.05 (Figure 1D). Notably, the grey module showed a positive correlation with RMD progression (r = 0.23, p = 0.02, Figure 1F), and its proteins with a higher DEPs (red ellipse) were primarily involved in innate immune responses and B-cell-related pathways (Figure 2A, 2B). 10 proteins were negatively associated with progression. DEPs with significant positive GS and AUC >0.6 included C1QL2, CD72, TNFRSF4, IL32, CXCL10, IL10, IL12B, and CLEC7A (Figure 2C). These proteins were also combined into B cell (C1QL2, CD72, IL32, TNFRSF4) and IFN protein scores (CXCL10, IL12B, CLEC7A, IL10), which enhanced predictive performance (Figure 2C). B cell scores were highest at baseline in progressors, as compared to after progression and in established SLE (Figure 2D). In contrast, IFN scores remained similarly elevated in all stages from before progression through to established disease, as compared with non-progressors or controls (Figure 2E). However, both were significantly distinct from non-progressors and healthy controls.
Conclusion: This study demonstrated the dysregulation of innate immune pathways among ANA-positive individuals even without clinical autoimmunity. A combination of IFN and B-cell-related immune aberration was most strongly associated with progression to clinical disease. These results suggest targeting one or both pathways in the highest-risk, biomarker-positive individuals might prevent SLE.
Significant protein identification and module construction from Weighted Gene Co-expression Network Analysis. (A) Venn diagram showing DEPs between each patient group. (B, C, D) volcano plots for DEPs in progressor vs. HC (B) and, non-progressor vs. HC (C), and progressors vs. non-progressors (D). (E) 193 selected proteins were constructed into seven modules. (F) Module trait correlation heatmap: column values show the correlation coefficient (Pearson’s, r) and p-value between the module and interested trait.
(A) GS value of proteins in the grey module, (B) the functional enrichment analysis of proteins in the grey module by GS. (C) ROC curve for biomarker performance of individual proteins or combined B cell and IFN protein scores. (D, E) bar plots comparing B cell and IFN scores across five groups: HC, At-risk non-progressors, At-risk progressors baseline, At-Risk progressors after progression, and established SLE.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: Porntip Intapiboon: None declared, Lucy M Carter: None declared, Jack Arnold: None declared, Md Yuzaiful Md Yusof Alumis, Roche and Novartis, Aurinia Pharmaceuticals, Edward M. Vital Roche, GSK, AstraZeneca, Aurinia Pharmaceuticals, Lilly and Novartis, Roche, AstraZeneca and Sandoz.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (