

Background: Antiphospholipid antibodies (aPL), which determine the diagnosis and pathophysiology of antiphospholipid syndrome (APS), fluctuate over time and can become undetectable. The significance of aPL-negativization is still unknown. Despite being crucial for aPL production in germinal centres, the study of B cell biology, including the molecular mechanisms regulating aPL production, is scarce in thrombotic primary APS (tPAPS).
Objectives: To characterize the immunophenotype and the molecular changes of circulating B cells regarding the aPL (positive/negativized) status.
Methods: Cross-sectional study of 62 prospectively recruited participants with tPAPS (Updated Sapporo Criteria) and 58 age- and sex-matched healthy donors (HD). Clinical and laboratory data were collected at study entry. Patients were grouped based on current aPL profile (lupus anticoagulant [LA]; IgM/IgG anti-cardiolipin [ACL], -ß2-glycoprotein [ß2GPI], -phosphatidylserine/prothrombin complex [PS/PT]; and IgG anti-Domain-1 of ß2GPI [D1- ß2GPI]) in: aPL+ (n=51), persistently positive for at least one aPL; aPL- (n=11), negative for all aPL (previously positive). The immunophenotype was analysed by flow cytometry using cryopreserved peripheral blood mononuclear cells. Then, using sorted B cells from 5 HC, 5 aPL- and 13 aPL+ patients, we sought to investigate the changes in gene expression leading the transition from an aPL+ to an aPL- state using single-cell RNA sequencing (scRNAseq).
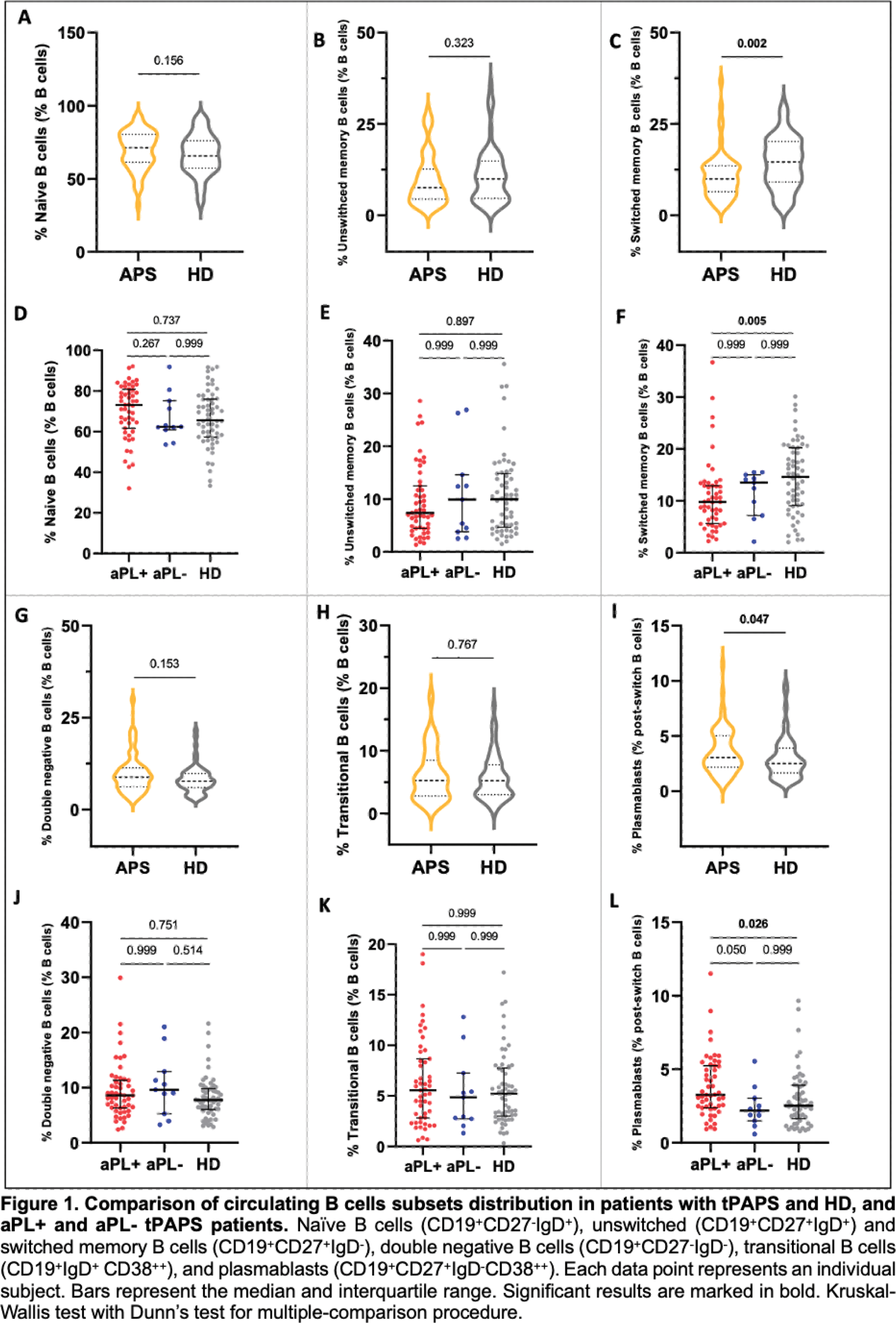
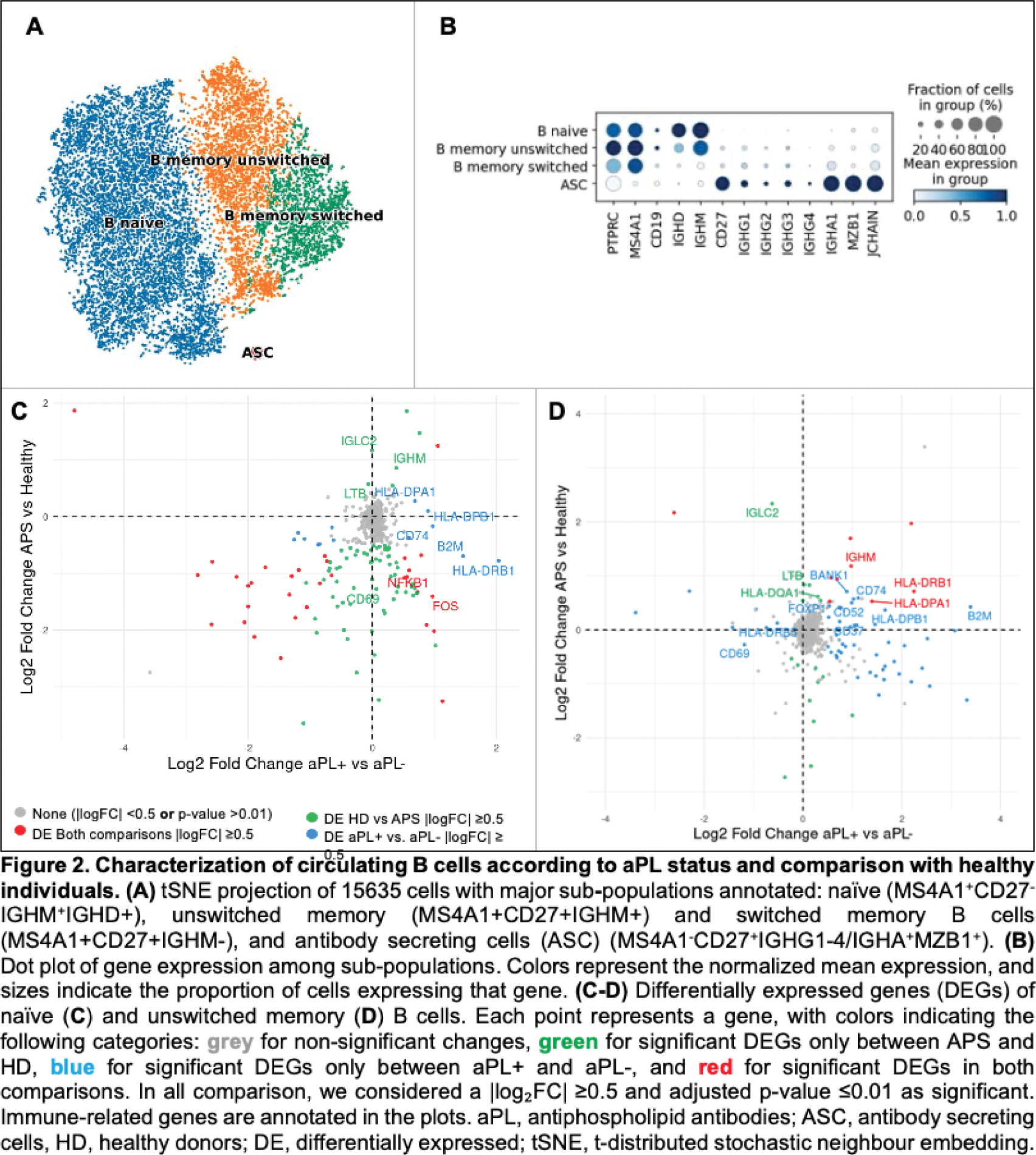
Results: Most patients were women (n=42, 67.7%), with a median (IQR) age at disease onset of 42.5 (29 - 52) years and a median follow-up of 5 (3 - 11) years. Arterial thrombosis was the most common first thrombotic manifestation (58.1%). Prevalence of criteria-aPL positivity decreased from diagnosis to follow-up: LA (53.2%; 33.9%); ACL (74.2%; 43.6%); ß2GPI (56.5%; 46.8%). Non-criteria-aPL anti-PS/PT and anti-D1-ß2GPI were currently present in 45.2% and 32.3% of patients, respectively. We observed aPL-negativization in 17.7% (n=11) of patients. The main demographic characteristics and treatment exposure did not differ between groups. The frequencies of circulating naive (Figure 1A and 1D), unswitched (Figure 1B and 1E), double negative (Figure 1G and 1J) and transitional (Figure 1H and 1K) B cells were similarly distributed between tPAPS patients and HD. Switched memory B cells (Figure 1C and 1F) were decreased and plasmablasts (Figure 1I and 1L) were increased in tPAPS compared to HD; these differences were observed between aPL+ patients and HD. To investigate the molecular alterations of B cells across different patient groups, we analysed their transcriptome using scRNAseq. This analysis allowed us to identify distinct B cell sub-populations (Figue 2A) and, using well-established phenotypic markers, classify cells as naïve, unswitched memory B cells, switched memory B cells and antibody-secreting cells (Figure 2B). Differential expression analyses were performed on the three main B cell subpopulations to compare the transcriptomic profiles between patients with tPAPS and HD, and between aPL+ and aPL- patients. Most differentially expressed genes (DEGs) were found in IGHM+ naive and unswitched memory B cells (Figure 2C and 2D). In naïve B cells, aPL+ patients have an upregulation of genes associated with immune activation, including CD74, B2M, and HLA-genes, in comparison to aPL- patients (Figure 2C). A similar transcriptional profile was observed in the unswitched memory B cells, where both tPAPS vs HD and aPL+ vs aPL- comparisons revealed significant gene expression changes, namely the upregulation of genes associated to adaptive cell activation (Figure 2D). Finally, switched memory B cells showed no significant DEGs.
Conclusion: Our analysis shows that the frequency of most circulating B cell populations is unchanged between tPAPS patients and HD and among different aPL status. However, the balance between the altered frequency of switched memory B cells and plasma cells in tPAPS might indicate ongoing antibody production. The upregulation of genes associated with immune activation in naïve and unswitched memory B cells points to a greater magnitude of adaptive immune responses in aPL+ patients, consistent with greater production of autoantibodies, even though their frequency is not changed. Therefore, it is critical to go beyond the analysis of the frequencies of distinct cell populations and address changes in the transcriptional program of those cells that can correlate with heightened autoimmune function.
REFERENCES: NIL.
Acknowledgements: We thank all participants for their contribution. This work is funded by National Funds through the Portuguese Funding agency FCT – Fundação para a Ciência e Tecnologia, I.P.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (