

Background: Calcium pyrophosphate deposition (CPPD) in joints is one of the most common causes of chronic arthropathy in individuals over 60 years of age. Historically, diagnosis was based on clinical presentation, evidence of crystals in synovial fluid analysis or typical radiographic findings. In recent years, ultrasound (US) has gained a central role in the diagnostic process. The OMERACT group has defined the elementary US lesions of CPPD and developed a scoring system for the extent of the deposition in the knees and wrists. Both elementary lesions and the scoring system have been thoroughly tested and validated for use in research and clinical practice. The 2023 ACR/EULAR Classification Criteria for CPPD Disease and the 2023 EULAR recommendations for the use of imaging in crystal-induced arthritis have endorsed the role of US in the diagnostic process of CPPD. Despite this progress, US remains an operator-dependent technique, and its effective application in CPPD diagnosis requires specialized training and expertise to minimize misinterpretations and diagnostic errors.
Objectives: The aim of this study is to develop an Artificial Intelligence (AI) tool for the identification and scoring of CPPD in the knee menisci.
Methods: The CLAIM study is an international OMERACT initiative involving the US CPPD and gout subtask groups, aiming to develop an AI algorithm for the identification and scoring of the elementary US lesions in gout and CPPD. Briefly, members of both OMERACT groups were asked to contribute with 108 high quality US images of CPPD (at triangular fibrocartilage of the wrist, hyaline cartilage and menisci of the knee) and 72 images of gout (double contour sign and tophi at any site), divided equally by degree (0-3) for the development of the scoring algorithm. In this abstract we present the preliminary results of the first deep learning approach for the development of the algorithm in CPPD patients at the menisci level. US images of the menisci, with any degree of deposition (from 0 to 3) as assigned by the participants and validated also by the steering committee, were included in this analysis. The region of interest, traced manually with dedicated software (3DSlicer software v5.6), included only the meniscus. For this first attempt, a transfer learning approach was employed using convolutional neural network (CNN) models pretrained on the ImageNet dataset. The models were fine-tuned with the training and validation data. Hyperparameters were systematically tested to identify the optimal CNN architecture and settings. To increase variability, the following random augmentations were applied during training: rotation (maximum ±20°), height shift (10%), width shift (10%), zoom range (10%), and horizontal flipping. Sample weighting correction was applied during training to address class imbalance. Each model was trained for a maximum of 100 epochs, with early stopping triggered if the validation loss did not improve after 10 consecutive epochs. Model performance was compared based on accuracy in classifying the validation set. Additional metrics - precision (correct classifications per predicted class), recall (correct classifications per actual class), and F1-score (harmonic mean of precision and recall) - were calculated for each grading class for the best-performing model. All metrics ranged from 0 to 1. All routines for image preprocessing and model implementation were conducted in Python v3.9, utilizing the OpenCV library v4.8 and the TensorFlow Keras framework v2.14.
Results: 19 rheumatologists from 10 countries (3 continents, Europe, Oceania and America) contributed with images in this first step. The final dataset consisted of 446 images with an unbalanced distribution among grades: 39 images (9%) for grade 0, 80 (18%) for grade 1, 215 (48%) for grade 2, and 112 (25%) for grade 3. The dataset was randomly split into training and validation sets (80% and 20%, respectively), yielding 356 and 90 samples while preserving the grading group distribution. The highest classification accuracy (CPPD yes/no) achieved on the validation set was 0.69. The overall performance for grading classes in the validation dataset is presented in Table 1. The precision of the deep learning model in correctly identifying true cases for each grading type was comparable for grades 0, 2, and 3 (ranging from 0.71 to 0.75) but lower for grade 1 (0.53). The confusion matrix displays the number of cases classified into each grading category within the validation dataset (Figure 1).
| precision | recall | f1-score | |
|---|---|---|---|
| grade 0 | 0.71 | 0.63 | 0.67 |
| grade 1 | 0.53 | 0.63 | 0.57 |
| grade 2 | 0.73 | 0.74 | 0.74 |
| grade 3 | 0.75 | 0.65 | 0.70 |
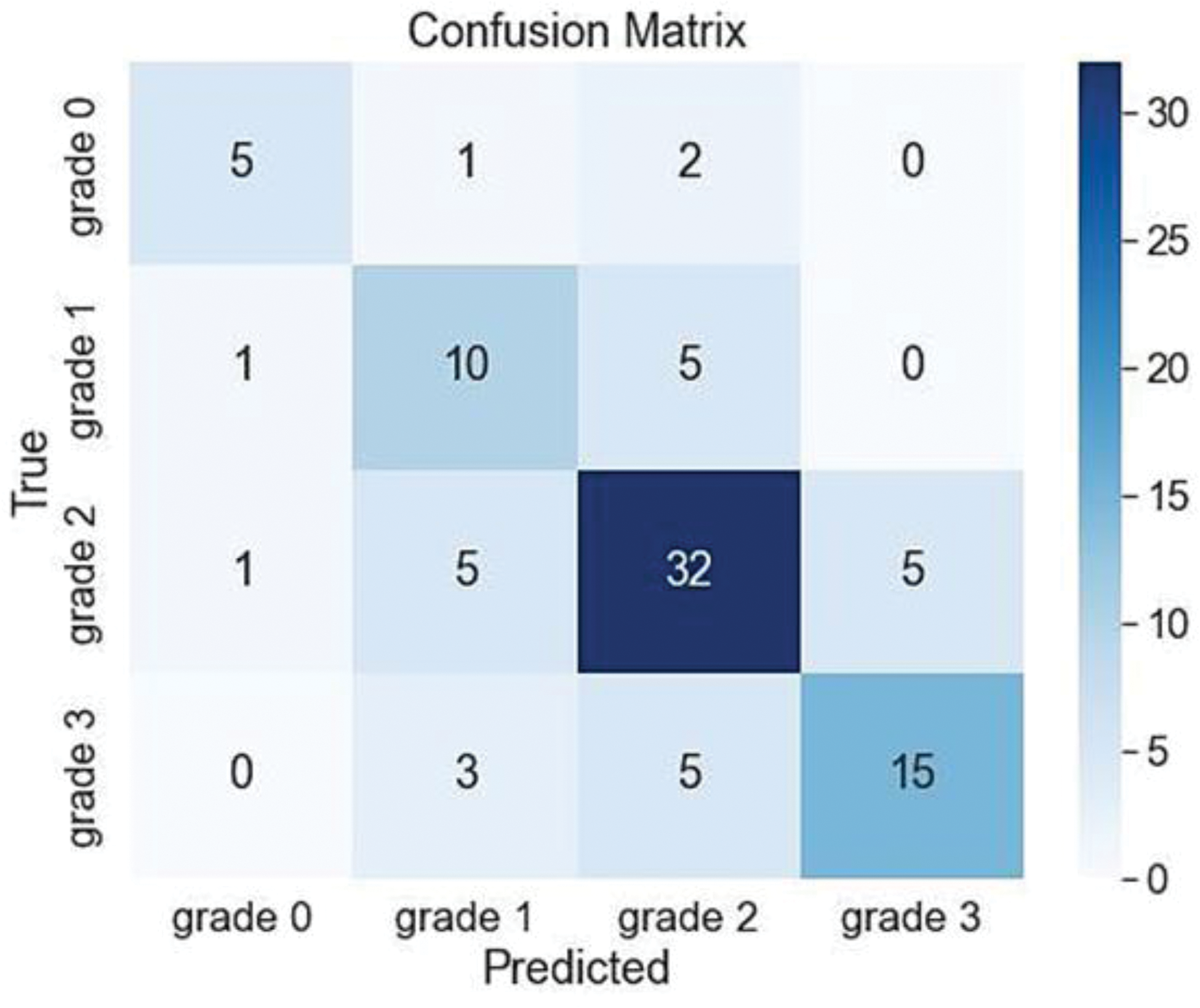
Conclusion: In conclusion, as the overall accuracy of the model falls slightly below the ‘good’ threshold level (0.69), the performance is insufficient for practical clinical application. This limitation may be partially attributed to the small dataset size, which is likely inadequate for training deep learning models effectively, even with fine-tuning of pretrained network architectures. Future work will prioritize increasing the dataset size, ensuring data balance, accounting for cross-validation procedure and exploring alternative approaches such as semantic segmentation of the cropped images.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: Georgios Filippou: None declared, Daniele Cirillo: None declared, Tito Bassani: None declared, Silvia Sirotti: None declared, Salvatore Gitto: None declared, Antonella Adinolfi Janssen, Janssen, Edoardo Cipolletta IBSA, Novartis, Horizion Therapeutics, Luis Coronel: None declared, Mario Diaz: None declared, Andrea Di Matteo Janssen, Emilio Filippucci: None declared, Hilde Berner Hammer Abbvie, UCB, Novartis, Lilly, Daryl MacCarter: None declared, Ingrid Möller Bristol-Myers Squibb, Pfizer, Johnson & Johnson, AbbVie, Gebro, Esperanza Naredo: None declared, Francesco Porta Laborest, IBSA, Amgen, Otto Martin Olivas Vergara: None declared, Garifallia Sakellariou: None declared, Wolfgang Schmidt: None declared, Ouidade Aitisha Tabesh: None declared, Giorgio Tamborrini: None declared, Plamen Todorov: None declared, Marta Arese: None declared, Rodolfo Fabbri: None declared, Alessandro Lucia: None declared, Antonio Varvaro: None declared, Piercarlo Sarzi-Puttini: None declared, Maria-Antonietta D’Agostino Amgen, MSD, BMS, Astrazeneca, GSK, Galapagos, Sanofi, Novartis, J&J, Abbvie, UCB, Pfizer, Alfasigma, Eli Lilly, Amgen, MSD, BMS, Astrazeneca, GSK, Galapagos, Sanofi, Novartis, J&J, Abbvie, UCB, Pfizer, Alfasigma, Eli Lilly, Peter Mandl AbbVie, Novartis, Janssen Sobi, AbbVie, Alfasigma, Novartis, Sobi, Carlos Pineda: None declared, Helen Keen: None declared, Luca Maria Sconfienza Esaote SPA, Samsung Medison, IBSA, Fidia Farmaceutici, Esaote SPA, Lene Terslev Novartis, UCB, Janssen, GE Healthcare.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (