

Background: Giant Cell Arteritis (GCA) is a systemic autoimmune disease affecting large and medium-sized arteries, with complications like vision loss if undiagnosed. Ultrasound (US) is a key diagnostic tool, but image interpretation expertise remains scarce, while US machines are widely available. A web-accessible artificial intelligence (AI), that physicians globally can upload images to, could assist in image classification, thereby simplifying the examination process and enhancing disease detection.
Objectives: This work investigates the application of supervised deep learning to develop an AI model that assists in US image classification for the presence or absence of GCA-typical lesions.
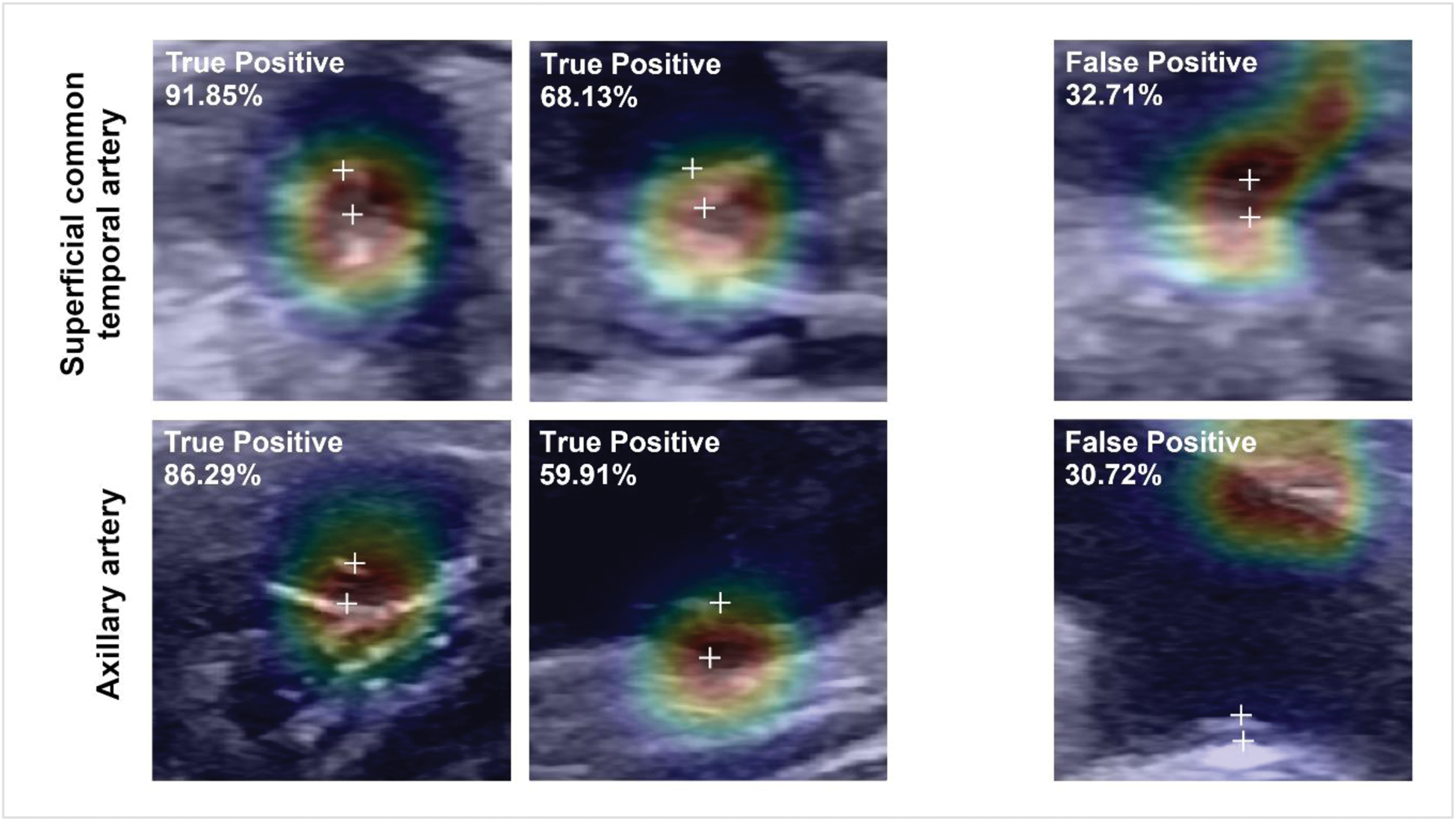
Methods: Our dataset includes 3,800 US images (38% displaying GCA-features) from 244 patients. In adequate representation of potential abnormalities [1], images were cropped to 224×224 pixels to match the ResNet input size. Using a ResNet-18-based architecture, we developed an US image classification model incorporating artery type (e.g., temporal, axillary) as meta-information. Artery-specific features were integrated by the model to enhance the detection of arterial wall thickening in US images, a key indicator of GCA [2]. Performance metrics, including F1-score (indicating the strength of true-positives identification and false-positives minimization) and area under the ROC curve (AUROC), were compared across three different model configurations (Artery-Specific Model, Artery Fusion Model and Combined Model) to select the most appropriate. Model interpretability and reliability were evaluated using Class Activation Maps and Monte Carlo Batch Normalization for uncertainty quantification.
Results: An affordable and efficient artery-fusion deep learning model was developed that is built to receive an ultrasound image and artery type information as input. Outperforming both other model configurations, the Artery Fusion Model achieved a superior diagnostic performance, precisely an F1-score of 81.12% and AUROC of 0.94 for the axillary artery, and an F1-score of 73.99% and AUROC of 0.87 for the superficial common temporal artery. Smaller branches of the superficial temporal artery demonstrated lower performance, reflecting inherent diagnostic challenges.
Conclusion: To date, the model developed in this project represents the world-leading artificial intelligence in GCA ultrasound image classification. Future work will focus on expanding datasets and incorporating multi-center validation to optimize detection in smaller arteries and enhance model generalizability. Consecutively, setup of a publicly accessible website hosting the AI-based assistant is envisioned in order to facilitate classification of uploaded ultrasound images and help to diagnose GCA early. This initiative holds great potential for the rapid and accurate diagnosis of GCA, prevention of ischemic complications through timely intervention and therewith high individual impact; particularly in resource-limited settings and regions.
REFERENCES: [1] Schäfer VS, Chrysidis S, Dejaco C, […], Bauer CJ. Exploring the Limit of Image Resolution for Human Expert Classification of Vascular Ultrasound Images in Giant Cell Arteritis and Healthy Subjects: The GCA-US-AI Project [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9).
[2] Schäfer VS, Juche A., Ramiro S., et al.: Ultrasound cut-off values for intima-media thickness of temporal, facial and axillary arteries in giant cell arteritis. Rheumatology 56(9), 1479–1483 (9 2017).
Illustration of exemplary Grad-Class Activation Maps for superficial common temporal artery (top) and axillary artery (bottom) from the Artery Fusion Model with corresponding label (upper left corner of each ultrasound image) and classification certainty (in percent). [True Positive= Images correctly classified as abnormal by the AI]. [False Positive= Images falsely classified as abnormal by the AI]
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (