

Background: Fibromyalgia, characterised by chronic pain and fatigue constitutes a major clinical problem in Europe and elsewhere. Hypermobile conditions, including Ehlers-Danlos syndrome and joint hypermobility syndrome, encompass a broad spectrum of heterogenous clinical entities characterised by excessive range of joint movements and variable extra-articular features. The etiological basis of these conditions is thought to relate to abnormal extra cellular matrix (e.g. collagen) assembly, which impairs the structural integrity of connective tissue. All rheumatologists will recognise a clinical phenotype characterised by fibromyalgia, neuropsychiatric features ( eg anxiety, bipolar disorder, ADHD, autism), hypermobility, and autonomic dysfunction ( eg POTS). We have also recently described a strong association between ‘Long COVID’ (in which condition chronic pain and fatigue are major features) and hypermobility [1]. However, the pathologic mechanisms underlying these clinical associations remain both debated and incompletely understood [2].
Objectives: In this study we applied a transcriptomic approach to explore gene expression in fibromyalgia, at baseline, and after a controlled experimental inflammatory challenge. Our hypothesis is that hypermobility is constitutionally associated with potentially deranged immunological function and following an inflammatory insult, a range of different pathways are triggered that result in neuropsychiatric, rheumatological and cardiovascular sequelae.
Methods: We tested for the effects of hypermobility on gene expression, both at baseline and following an inflammatory challenge, by analysing existing transcriptomic data gathered from healthy subjects and patients with fibromyalgia and ME/CFS (ISRCTN78820481) using total RNA sequencing of blood samples (Illumina NextSeq 500). From each participant, two blood samples were taken on separate occasions: one after a saline injection and the other after an inflammatory challenge, typhoid vaccination, in a double-blind, randomised, cross-over design. Participants were assigned to hypermobile or non-hypermobile group according to Criterion 1 of the 2017 for Hypermobile Ehlers-Danlos Syndrome (hEDS) diagnostic criteria. Differential gene expression analysis comparing hypermobile and non-hypermobile participants was performed using EdgeR software in R studio. Baseline differences were determined by comparing post-saline samples between groups. Differential responses to the inflammatory challenge were detected by comparison of post-typhoid samples between groups. Gene ontology on differentially expressed genes was performed using DAVID’s Functional Annotation Clustering tool.
Results: Gene ontology analysis of differentially expressed genes (hypermobile versus non-hypermobile) at baseline were found to associate with six diseases (AIDS, leiomyoma, cardiomegaly, eczema, atopic dermatitis and SLE). After inflammatory challenge compared to baseline, the hypermobile group displayed a gene expression profile associated with 27 diseases including substance use disorders, bipolar disorder, autism, ADHD and Parkinson’s disease, myocardial infarction and atrial fibrillation. (Figure 1. Gene ontology analysis in hypermobile versus non- hypermobile individuals following an inflammatory challenge.).
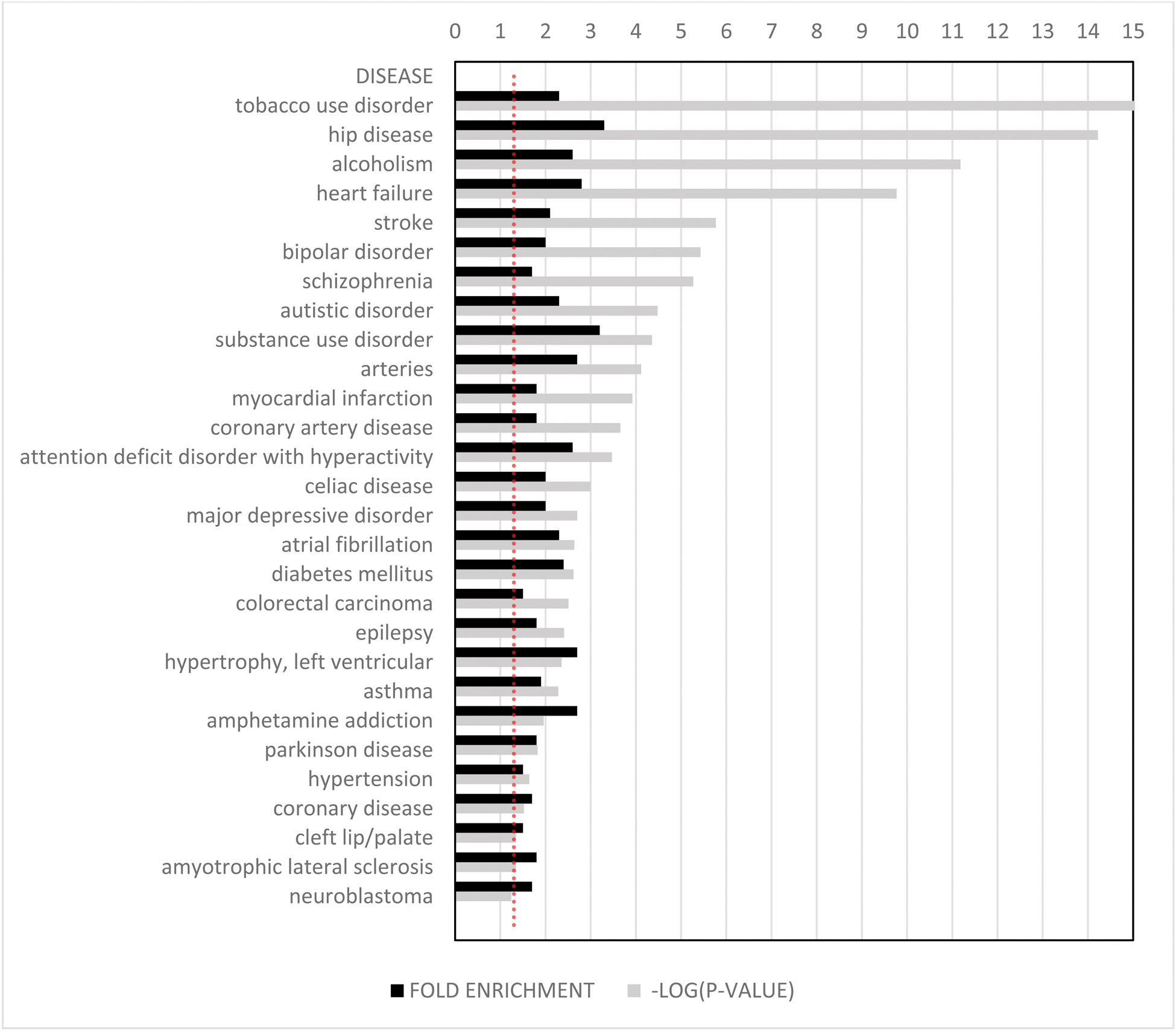
Conclusion: Here, we note at baseline, multiple associations with hypermobility and immunological disease, both immunodeficiency and autoimmunity, including SLE and atopic dermatitis, which suggests impaired immunological regulation in hypermobile patients. After inflammatory challenge there were associations with a large number of cardiovascular and neuropsychiatric conditions, notably ADHD and bipolar disorder. This suggests a two-factor model whereby hypermobility is indeed constitutionally associated with deranged immunological function. Following an inflammatory insult, neuropsychiatric and cardiovascular sequelae may occur. This model potentially explains the common association of fibromyalgia/CFS and mental health disorders with what may be an acute, often infective, inflammatory precipitant, or in some instances a more chronic auto-immune stimulus. Further research is clearly needed to delineate the nature of such mechanistic links, with a view to identifying potential therapeutic targets, and/or developing a personalized medicine approach which may mitigate the risk of developing chronic sequelae after say, COVID or other viral infection, in vulnerable individuals.
REFERENCES: [1] Jessica A Eccles, Dorina Cadar, Lisa Quadt, Alan J Hakim, Nicholas Gall, Covid Symptom Survey Biobank Consortium, Vicky Bowyer, Nathan Cheetham, Claire J Steves, Hugo D Critchley, Kevin A Davies - Is joint hypermobility linked to self-reported non-recovery from COVID-19? Case–control evidence from the British COVID Symptom Study Biobank: BMJ Public Health 2024; 2:e000478.
[2] Jessica A Eccles, Beth Thompson, Kristy Themelis, Marisa L Amato, Robyn Stocks, Amy Pound, Anna-Marie Jones, Zdenka Cipinova, Lorraine Shah-Goodwin, Jean Timeyin, Charlotte R Thompson, Thomas Batty, Neil A Harrison, Hugo D Critchley, Kevin A Davies. Beyond bones: the relevance of variants of connective tissue (hypermobility) to fibromyalgia, ME/CFS and controversies surrounding diagnostic classification: an observational study. Clin Med (Lond ) 2021;21:53–8.
Acknowledgements: Versus Arthritis.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (