

Background: Childhood-onset systemic lupus erythematosus (cSLE) is a chronic autoimmune rheumatic disease with frequent onset at the time of puberty and strong female bias (4.5:1). This indicates that sex determinants may play a role in this immunological dimorphism associated with cSLE risk. Sex hormones signal via their receptors on immune cells, with impact on their phenotype, metabolism and function. The extent to which sex hormones and chromosomes influence immune cells, and their effect on autoimmune susceptibility is not fully explored.
Objectives: This study aimed to investigate immune transcriptomic changes induced by sex hormones and different sex chromosomal backgrounds in transgender adolescents and their association with cSLE.
Methods: We performed peripheral blood mononuclear cell (PBMC) RNA sequencing analysis on post-pubertal cisgender adolescents with cSLE (n=74, all cis-female, mean age/disease duration: 19/6.5 years) and healthy controls (HCs, n=20, all cis-female, mean age: 18), as well as adolescent transgender individuals (trans-male, XX TM; trans-female, XY TF) undergoing gender-affirming hormone therapy (GAHT) across 3 timepoints; pre-treatment (n=10, 5 XX TM, mean age: 15.9, 5 XY TF, mean age: 16.6), on gonadotrophin-releasing hormone agonist (GnRHa, puberty blocker) (n=12, 7 XX TM, mean age: 17.1, 5 XY TF, mean age: 18.3) and on GAHT, TM on testosterone and TF on oestradiol (n=10, 5 XX TM, mean age: 18.2 5 XY TF, mean age: 19.4). Rstudio was used to identify the significantly differentially expressed genes (DEGs, DEseq2) between groups and timepoints (log fold change (FC)>1.5, unadjusted P-value <0.01 or <0.05). We performed pathway enrichment analysis via Metascape, clustering and PCA analysis through Clustvis and normalised gene count analysis with Graphpad Prism.
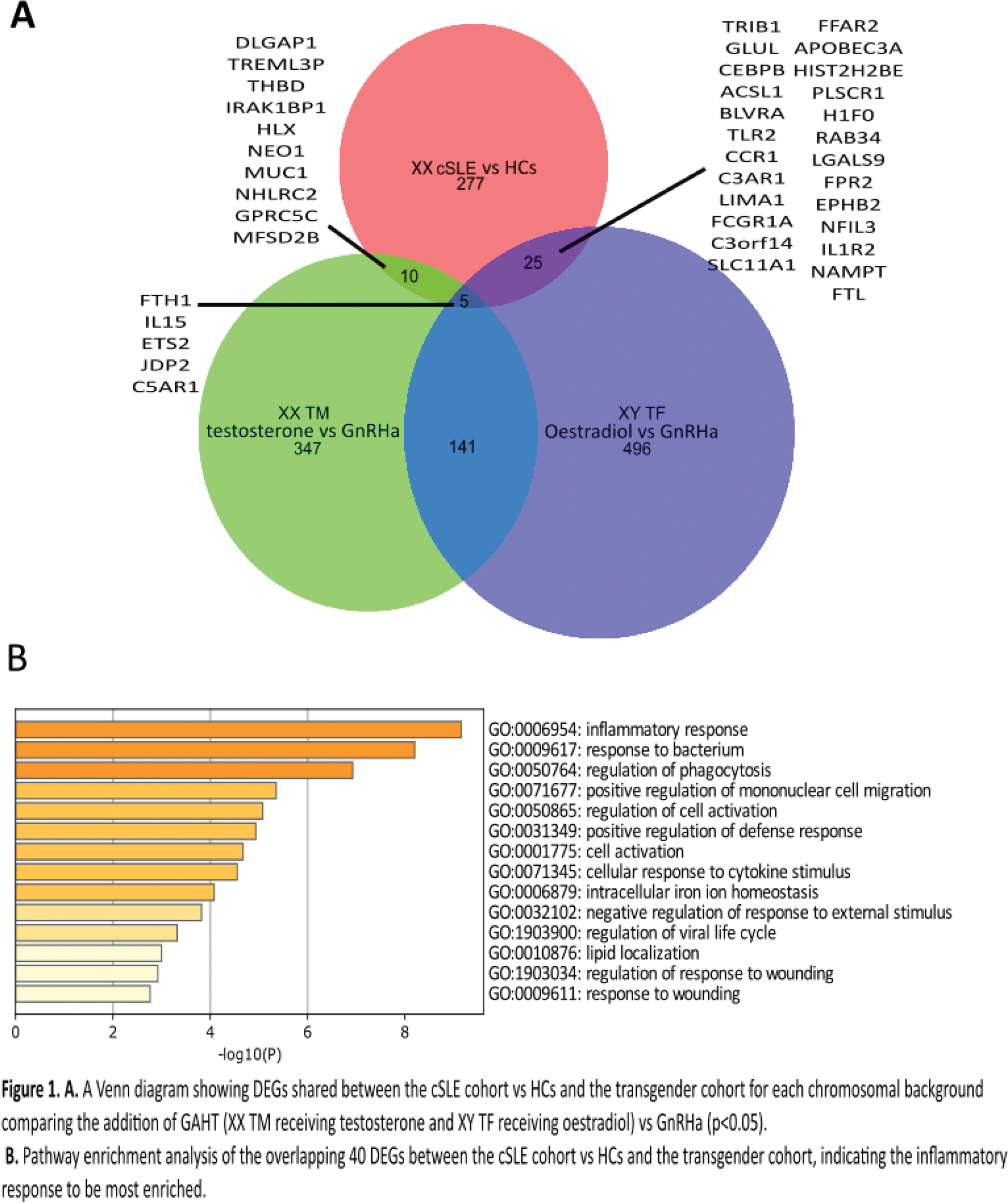
Results: We identified a total of 40 DEGs (FC>1.5, P<0.05) in the cis-female cSLE cohort vs. HCs (defining a post-pubertal cSLE signature) that overlapped with those associated with GAHT in the transgender cohort (Figure 1A). Specifically, in the XY TF group (on oestradiol vs. GnRHa), there were 24 upregulated and 1 downregulated DEGs, including TLR2 (p= 0.0077), C3AR1 (p= 0.040) and IL1R2 (p= 0.048) which overlapped with the cSLE signature. Similarly, in the XX TM group (on testosterone vs. GnRHa), there were 6 upregulated and 4 downregulated DEGs that overlapped, including IL15 (p= 0.019) and NEO1 (p= 0.023). Pathway enrichment analysis revealed that the majority of these sex hormone driven and cSLE overlapping genes were involved in inflammatory response processes, cell migration, activation and lipid regulation (Figure 1B). Across the transgender cohort analysis, we also discovered 63 X-linked DEGs (P <0.05) in the GAHT vs. GnRHa groups across both sex chromosomal backgrounds, however, these did not overlap with the cSLE signature, suggesting a less profound impact of sex hormones on X-linked transcripts in relation to cSLE susceptibility. Despite this, the X-linked non-coding RNA X-inactive specific transcript ( XIST ) and the XIST antisense non-coding RNA TSIX , were both significantly increased in cSLE vs. HCs (p=0.018 and 0.0007). XIST correlated negatively with cSLE interferon scores (p=0.0010, r=-0.38) and X-linked TLR7 expression (p=0.0004, r=-0.40), but TSIX did not, supporting a role for X chromosome inactivation in controlling the pathogenic mechanisms associated with cSLE in females.
Conclusion: This analysis demonstrated unique inflammatory transcriptomic changes (DEGs) induced by gender-affirming hormone therapy in transgender adolescents, which overlapped with those found in an adolescent cis-female cSLE cohort, as well as sex hormone-independent associations between X-linked XIST expression and inflammatory cSLE pathogenic mechanisms in adolescent cis-females. To our knowledge, this is the first exploration of the impact of sex hormones disaggregated from their corresponding sex chromosomal background in driving transcriptomic changes potentially relevant for understanding female cSLE pathogenesis.
REFERENCES: NIL.
Acknowledgements: Thank you to all the individuals who generously donated their blood and data to this study.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (