

Background: Sjögren’s disease (SjD) is a systemic autoimmune disorder characterized by a highly heterogeneous clinical and biological presentation. Identifying SjD subgroups with distinct pathobiological characteristics can enhance the understanding of pathogenetic mechanisms and support the development of targeted therapies tailored to the specific needs of each subgroup. Despite this potential, to date, only limited information exists on clustering SjD patients based on the characteristics of affected tissues, such as salivary glands.
Objectives: Our aims were (i) to identify potential subgroups of patients with SjD in an explorative study by proteome analysis of minor salivary gland (MSG) tissues, and (ii) to determine whether identified subgroups were reflected systemically at levels of PBMC transcriptomes and serum cytokine profiles.
Methods: Flash frozen MSG, PBMC and serum from 18 SjD patients, fulfilling the 2016 ACR/EULAR classification criteria [1] were analyzed, and their clinical data were collected. This included demographic, histological, imaging, functional and laboratory parameters, such as age, gender, focus score, number of germinal centers (GC), presence of lymphoepithelial lesions, salivary gland ultrasound scores (SGUS), Schirmer’s test, unstimulated salivary flow (USF) test, and presence of autoantibodies. Proteome analysis of the MSG tissues was performed by tandem mass spectrometry. Proteins were identified and quantified using the Spectronaut software. Principal component analysis (PCA) was performed based on log2-transformed Spectronaut intensities (SI). Interferon (IFN) signatures were calculated according to Gottenberg et al [2]. Bayes test was applied to identify differentially expressed proteins (log2 FC≥ 1, p-adj ≤0.05) between clusters. Pathway enrichment analysis for Reactome and KEGG pathways was performed on differentially expressed proteins using the STRING database, and an enrichment score ≥1. RNA sequencing was performed on RNA from PBMC, and PCA was performed based on the 500 most variable genes. Serum levels of IL6, IL8, CXCL10, CXCL13, CCL19, and BAFF were measured with multi-analyte immunoassays. Differences between clusters were analyzed using the Mann-Whitney U test or Fisher’s exact test.
Results: Proteome analysis detected 8192 proteins in MSG tissues. PCA identified three separate clusters of patients with SjD (Figure 1A). Patients from cluster 1 (n=3) had higher focus scores, a higher number of GC and a lower USF when compared to clusters 2 (n=9) and 3 (n=6). In addition, the percentage of patients testing positive in SGUS was higher in cluster 1. Patients from cluster 1 had higher IFN signatures, and higher expression of CXCL10 and CXCL13 in MSG. In addition, cluster 1 had a higher percentage of patients positive for rheumatoid factor (RF) and cryoglobulins (Figure 1B). Analysis of differentially expressed proteins between clusters showed that in cluster 1, 181 proteins were upregulated and 33 were downregulated compared to cluster 2, and 263 proteins were upregulated and 186 downregulated compared to cluster 3. In cluster 2, 250 proteins were upregulated and 41 were downregulated when compared to cluster 3. Pathway enrichment analysis identified IFN signaling, antigen presentation and T cell activation as key characteristics of proteins in cluster 1 vs clusters 2 and 3. In cluster 1, pathways related to innate immune system, neutrophil degranulation and amino acid metabolism were suppressed compared to cluster 3. In cluster 2, pathways related to RNA metabolism and rRNA processing were upregulated compared to cluster 3, whereas pathways related to lysosome and secretory and luminal granule proteins were suppressed. Transcriptome analysis of PBMC indicated that patients clustered differently based on PBMC transcriptomes compared to the MSG proteome, with patients from the three MSG proteome clusters being distributed in all clusters based on PBMC transcriptomes (Figure 1C). Additionally, serum levels of IL6, IL8, CXCL10, CXCL13, CCL19 and BAFF did not significantly differ in patient clusters based on MSG proteomes (Figure 1B).
A. PCA based on log2-transformed Spectronaut intensities (SI) of detected proteins in MSG tissues. B. Clinical characteristics and laboratory measures in patient subgroups as defined in A. C. PCA based on the 500 most variable genes detected in PBMC.
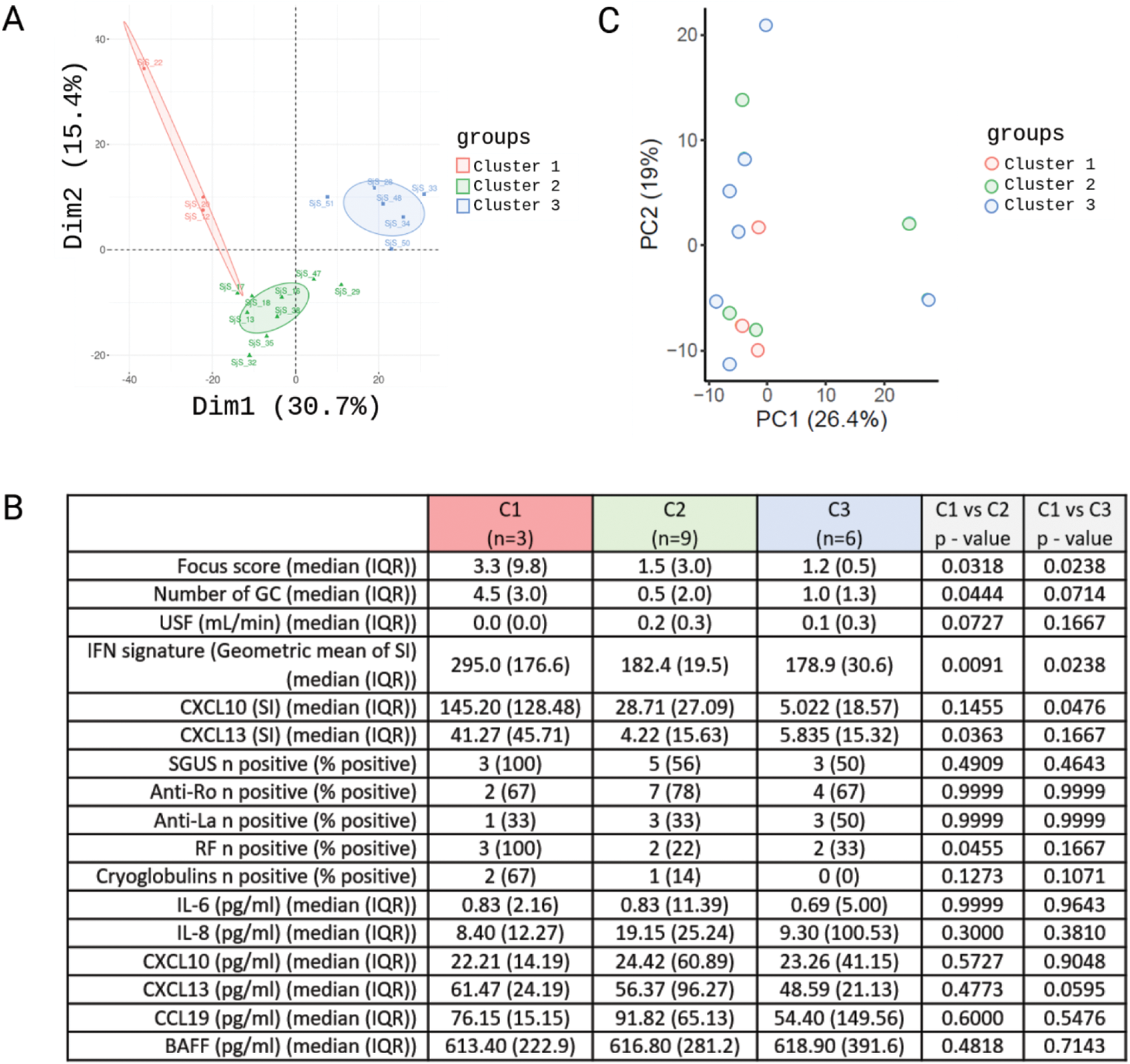
Conclusion: SjD patients in our cohort clustered in 3 subgroups based on MSG proteomes, with the first subgroup showing the most prominent glandular damage, high T cell activation and the highest IFN scores. Subgroups defined based on MSG proteomes were not reflected systemically by PBMC transcriptomes and serum cytokine levels suggesting that tissue-based analysis better reflects pathobiological activity.
REFERENCES: [1] Shiboski CH, Shiboski SC, Seror R, Criswell LA, Labetoulle M, Lietman TM, et al. 2016 American College of Rheumatology/European League Against Rheumatism Classification Criteria for Primary Sjögren’s Syndrome: A Consensus and Data‐Driven Methodology Involving Three International Patient Cohorts. Arthritis & Rheumatology. 2017;69(1):35-45.
[2] Gottenberg JE, Cagnard N, Lucchesi C, Letourneur F, Mistou S, Lazure T, et al. Activation of IFN pathways and plasmacytoid dendritic cell recruitment in target organs of primary Sjögren’s syndrome. Proc Natl Acad Sci U S A. 2006;103(8):2770-5.
Acknowledgements: NIL.
Disclosure of Interests: Neža Štucin: None declared, Katja Perdan Pirkmajer: None declared, Alojzija Hočevar: None declared, Sara Andrea Krättli: None declared, David Miguel Ferreira Francisco: None declared, Britta Maurer Lecturing fees from Boehringer Ingelheim, GaxoSmithKline, Novartis, Otsuka, and Merck Sharpe & Dohme, Is on an advisory board for Janssen and Boehringer, Consulting fees from Novartis, Boehringer Ingelheim, Janssen-Cilag, and GaxoSmithKline, Research grants from AbbVie, Protagen, and Novartis Biomedical Research, Rémy Bruggmann: None declared, Polona Zigon: None declared, Saša Čučnik: None declared, Kerstin Klein: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (