

Background: Giant cell arteritis (GCA) is one of large vessel vasculitis affecting mainly elderly patients, which causes headaches, pain in the jaw muscles, and visual loss. The tissues of temporal arteries in patients with GCA show characteristic pathological features including perivascular granuloma formation, lymphocyte infiltration, and elastic lamina destruction. We previously identified MMP12 as the most significantly upregulated molecule in vasculitis through serum proteome analysis and validation experiments. However, the role of MMP12 in the pathophysiology of GCA is still unclear.
Objectives: The objective of this study is to analyze the detailed pathological features of GCA through spatial transcriptome analysis using affected temporal artery biopsy specimens and to elucidate the contribution of MMP12 to the pathophysiology of GCA.
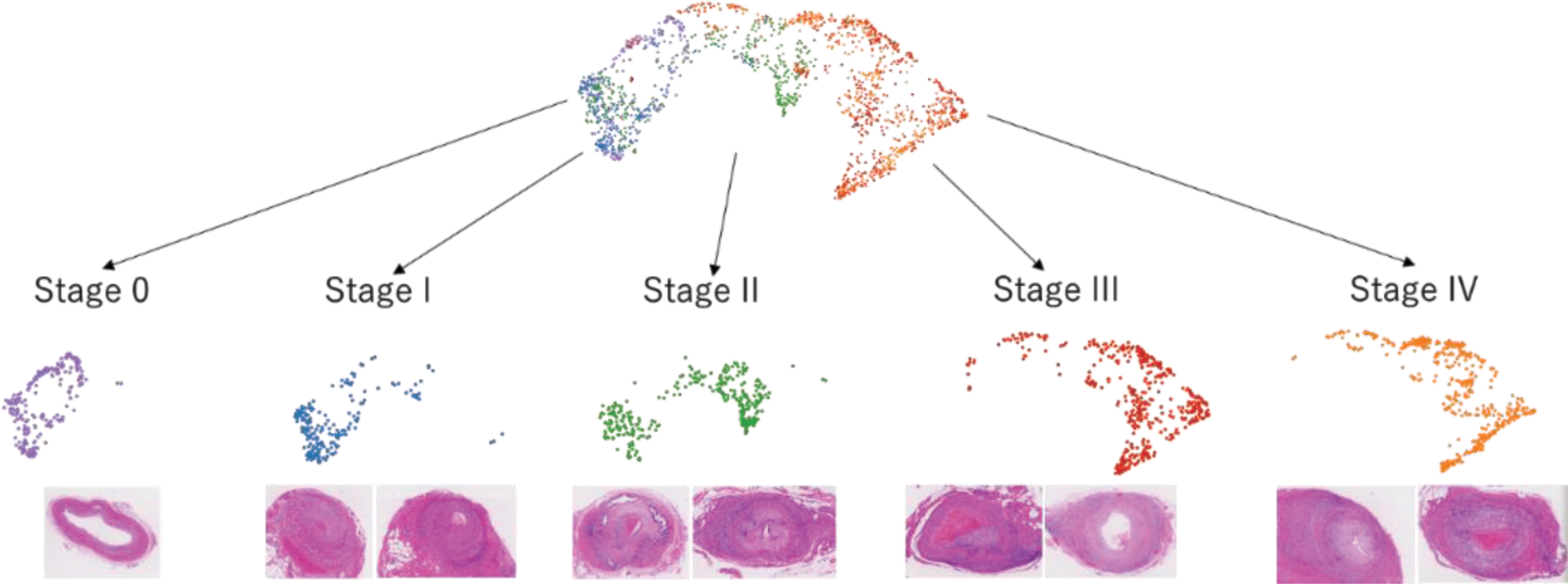
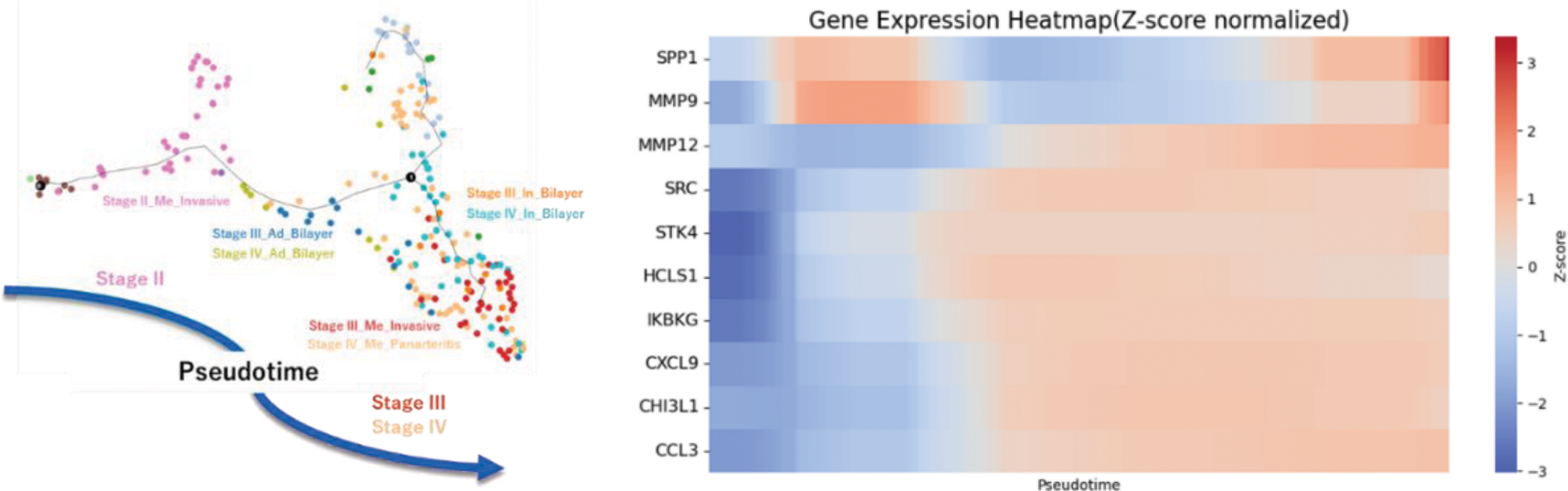
Methods: We performed spatial transcriptome analysis using 10x Visium on eight GCA biopsy samples. Each spot was histologically classified and annotated as macrophage, multinucleated giant cell (MGC), lymphoid, intima, media, or adventitia. We classified the lesions into Stages 0, I, II, III, and IV using the hypothetical model proposed by Hernández-Rodríguez J, et al [1] and analyzed stage progression on the UMAP of gene expression profiles for each spot. We then extracted macrophage and MGC spots, performed sub-clustering, and conducted trajectory analysis. The relationship between pseudotime and gene expression of proteins upregulated in the peripheral blood of patients with vasculitis was analyzed.
Results: The transition of UMAP clusters was aligned with histopathological stage classification (Figure 1). Pseudotime analysis of extracted macrophage and MGC correlated with histopathological progression from Stage II to Stage III/IV. Among the proteins whose gene expression were upregulated in the sera of patients with vasculitis, the top 10 expressed genes in lesional macrophages and MGCs in spatial transcriptome analyses were SPP1, MMP9, CHI3L1, HCLS1, CXCL9, STK4, SRC, CCL3, IKBKG, and MMP12. SPP1 and MMP9 are known to be involved in the pathogenesis of GCA [2, 3]. Whereas the Z-score of those molecules showed a bimodal increase in the early and late pseudotime, that of MMP12 exhibited broad and steady increase in the latter half of pseudotime (Figure 2). Notably, MMP12 exhibited the most significant change in expression across pseudotime, with a ratio of approximately 50 between its minimum and maximum values.
Conclusion: The spatial transcriptome analysis with pseudotime analysis revealed a distinctive pattern of gene expression of MMP12. Its broad and steady increase in gene expression profile, along with the largest upregulation ratio exceeding 50-fold across disease stages, suggests its role in the pathogenesis of GCA. This finding suggests that MMP12 is not only a novel, promising biomarker for GCA in monitoring disease activity and structural remodeling but also has a potential to be a treatment target.
REFERENCES: [1] Hernández-Rodríguez J, et al. Medicine 95(8): e2368, 2016.
[2] Watanabe R, et al. Circ Res. 2018;123:700-715.
[3] Prieto-González S, et al. RMD Open 2017;3: e000570.
UMAP cluster transition aligned with histopathological stage classification.
Pseudotime analysis of extracted macrophage and MGC correlated with histopathological stage progression from Stage II to Stage III/IV. The gene expression of MMP12 exhibited broad and steady increase in the latter half of pseudotime.
Acknowledgements: NIL.
Disclosure of Interests: Shinichi Onishi Chugai Pharmaceutical Co. Ltd., Hiroto Yoshida Chugai Pharmaceutical Co. Ltd., Mayu Magi Chugai Pharmaceutical Co. Ltd., Chie Kato Chugai Pharmaceutical Co. Ltd., Kotaro Matsumoto: None declared, Katsuya Suzuki: None declared, Masaru Takeshita: None declared, Junko Kuramoto: None declared, Masaki Yazawa: None declared, Tatsuya Kato: None declared, Sota Yamada: None declared, Eita Sasaki: None declared, Kenjiro Hanaoka: None declared, Mitsuhiro Akiyama: None declared, Yasushi Kondo: None declared, Jun Kikuchi: None declared, Yuko Kaneko Chugai Pharmaceutical Co. Ltd.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (