

Background: CD64, encoded by FCGR1A gene, is the most common Fc receptor expressed by macrophages, neutrophils, and eosinophils, with a primary role on antibody-mediated phagocytosis. We hereby describe the first clinical case of a patient with a pathogenetic variant in FCGR1A gene. Whole exome sequencing (WES) analysis revealed a second variant of unknown clinical significance (VUS) on DKC1 gene. Laboratory investigations were performed on proband and his relatives to elucidate the potential clinical implications of these genetic findings.
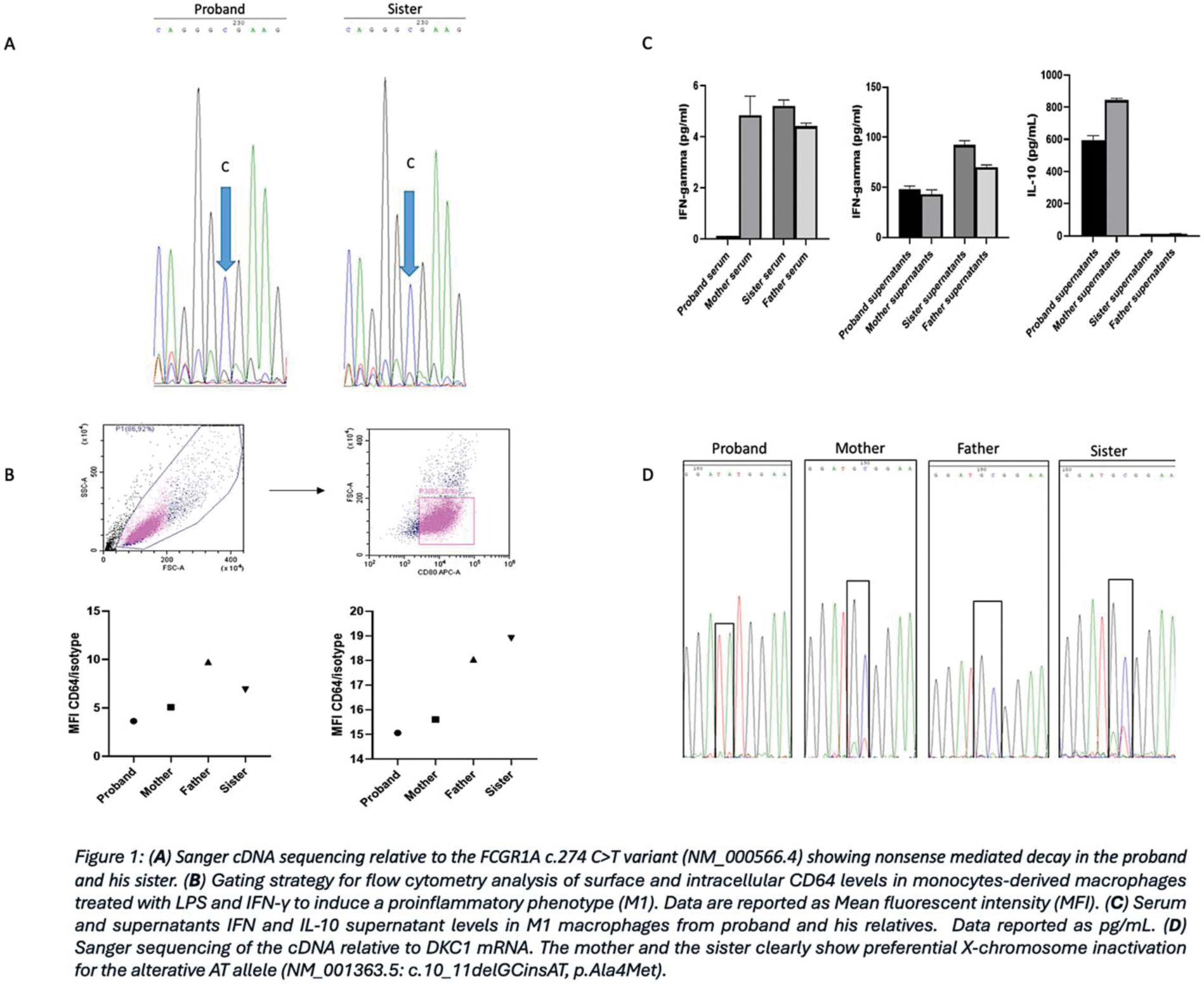
Case presentation: A 29-year-old male patient reported recurrent febrile episodes, pustulosis, oral aphthae and lymphadenopathy. Lymphocyte typing showed the presence of a double negative T cell population (25% of the total cells), and a relative reduction in NK cells (5%) and B cells (10%). Therapy with colchicine and IL-1 inhibitor proved ineffective. TNF inhibitor therapy resolved febrile episodes but no other symptoms, while isotretinoin and repeated courses of antibiotic therapies improved skin lesions. WES identified a rare variant in the FCGR1A gene classified as ‘pathogenetic’ in ClinVar ; the variant was maternally inherited in heterozygosity by the proband and his sister (Figure 1A). However, real-time PCR gene expression analysis showed a significant reduction in FCGR1A transcript levels in the proband, but not in other carriers. Furthermore, flow cytometry revealed a reduced expression of CD64 on the patient’s macrophages compared to those of his relatives (Figure 1B). Luminex analysis of 40 inflammatory mediators was performed on M1 macrophages supernatants and in the sera from all participants demonstrating a significant reduction of IFN-γ levels in the proband’s serum (Figure 1C). A second VUS variant was detected in the DKC1 gene both in the proband and his sister. Pathogenetic DKC1 variants have been previously associated with severe immunological defects, including decreased B and NK lymphocytes, and reduced IFN-γ serum levels. Sanger sequencing of the cDNA encoded by DKC1 (Figure 1D) clearly showed that the alternative allele (GC>AT) is downregulated in the proband’s sister and mother. Such a finding is supported by the skewed X-chromosome inactivation pattern, suggesting a possible pathogenetic role for the alternative AT allele in the proband, who lacks the compensatory wild-type allele.
Learning points for clinical practice: The pivotal role of the CD64 receptor in phagocytosis suggests that its reduction on macrophage surface may lead to an increased susceptibility to infections. It remains unclear whether the DKC1 variant is associated with the reduction in INF-γ level of the patient’s serum. However, this specific cytokine profile, combined with the FCGR1A pathogenetic variant, may contribute to the dysfunction of CD64-related immunological pathways, leading to dysregulated skin inflammation.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (