

Background: Around 70% of Systemic Lupus Erythematosus (SLE) patients follow a relapsing-remitting pattern of disease, with unpredictable flares of disease activity followed by variable periods of disease quiescence. CD4 + T cell subsets have been shown to play an important role in driving the autoantibody production that causes flares in SLE, however the precise T cell changes that accompany flares are unknown.
Objectives: To characterize the antigen-experienced memory T cell compartment at various phases of disease to gain insight into the T cells changes that accompany flares.
Methods: CITE-seq was used to assess the transcriptomic profiles of CD4 + memory T cells in flaring (change in the clinical SLEDAI-2K > 0 in the last month prompting a change in therapy) and quiescent (clinical SLEDAI = 0 for at least a year, prednisone dose < 10) SLE patients. CD4 + memory T cells were isolated from previously archived PBMCs, stained with oligo-conjugated antibodies against surface proteins, and then partitioned, barcoded, and sequenced. Samples from 15 distinct patients at two separate clinical visits spaced at least one year apart were examined. TCR sequencing was performed to assess clonotype expansion/contraction. The longitudinal nature of our data permitted examination of transcriptional changes both between and within patients.
Results: Integration of the gene and surface protein expression data led to identification of 23 unique immune populations [Figure1.A]. Samples from flaring patients were more enriched for T follicular helper (Tfh), T peripheral helper (Tph), and Th1 cells. Conversely, quiescent patients were more enriched for central memory T cells (TCMs), specifically TCM1/2/6, compared to flaring patients (p < 0.05) [Figure1.B&C]. There was no difference in the proportion of exhausted Treg cells between flaring and quiescent patients at baseline suggesting that exhaustion of this subset is a consistent feature of SLE, potentially contributing to an inherent level of immune dysregulation regardless of clinical flare status. Differential gene expression analyses at baseline showed a high prevalence of IFN-induced genes in most identified cell subsets within the genes that were upregulated in flaring compared to quiescent patients, with a few exceptions (e.g., Th1). TCR repertoire analyses of samples at baseline revealed a higher proportion of expanded clonotypes in various clusters, such as Th17 cells, in flaring patients [Figure 2.A], which was not seen in quiescent patients [Figure 2.B]. Similarly, clonal overlap among subsets was more pronounced in the flaring patient samples [Figure 2.C] than in the quiescent samples [Figure 2.D]. This overlap indicates that these T cells have shared antigen specificity, suggesting cell plasticity or differentiation from a common precursor – which will be investigated by trajectory pathway analyses.
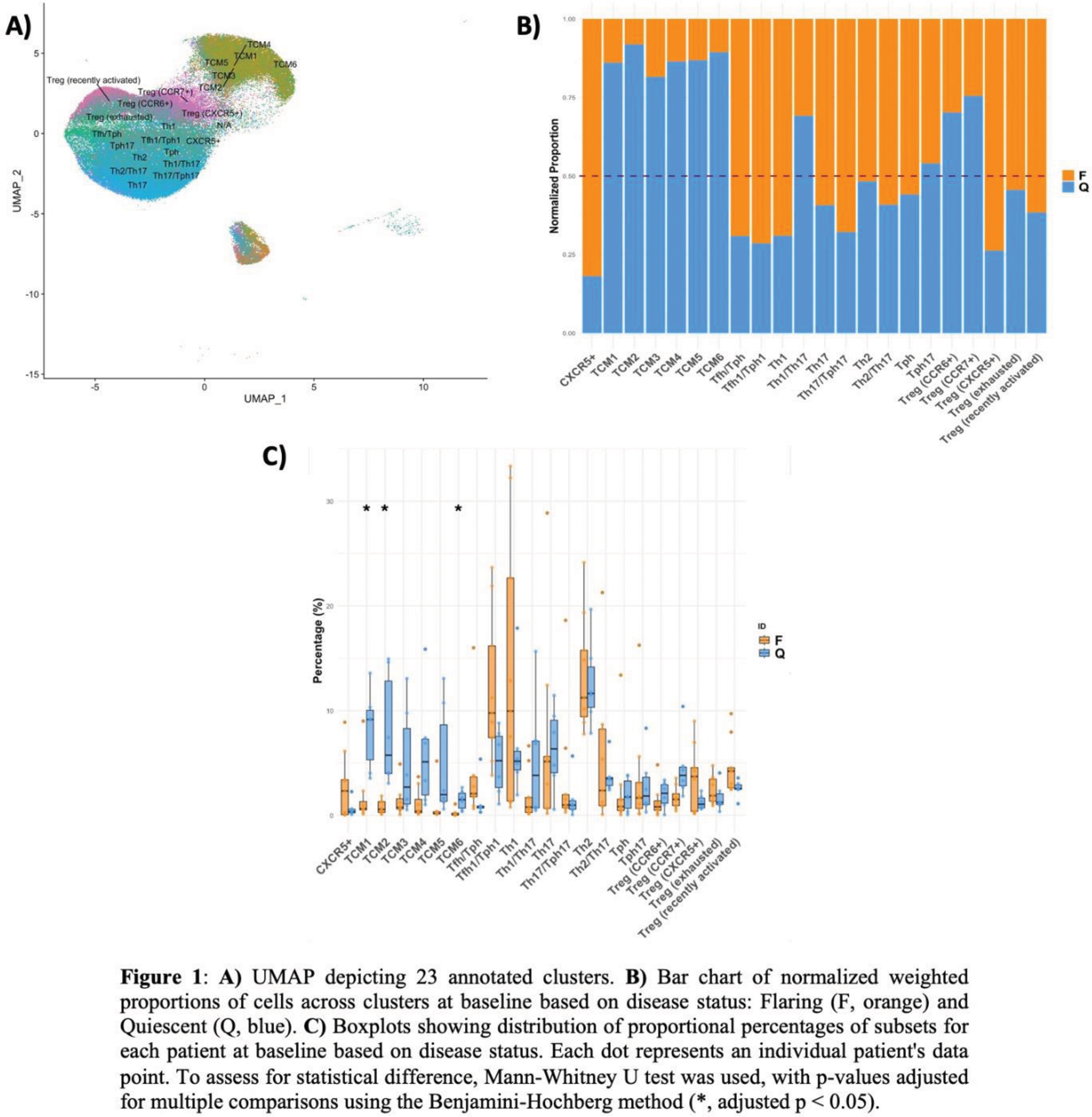
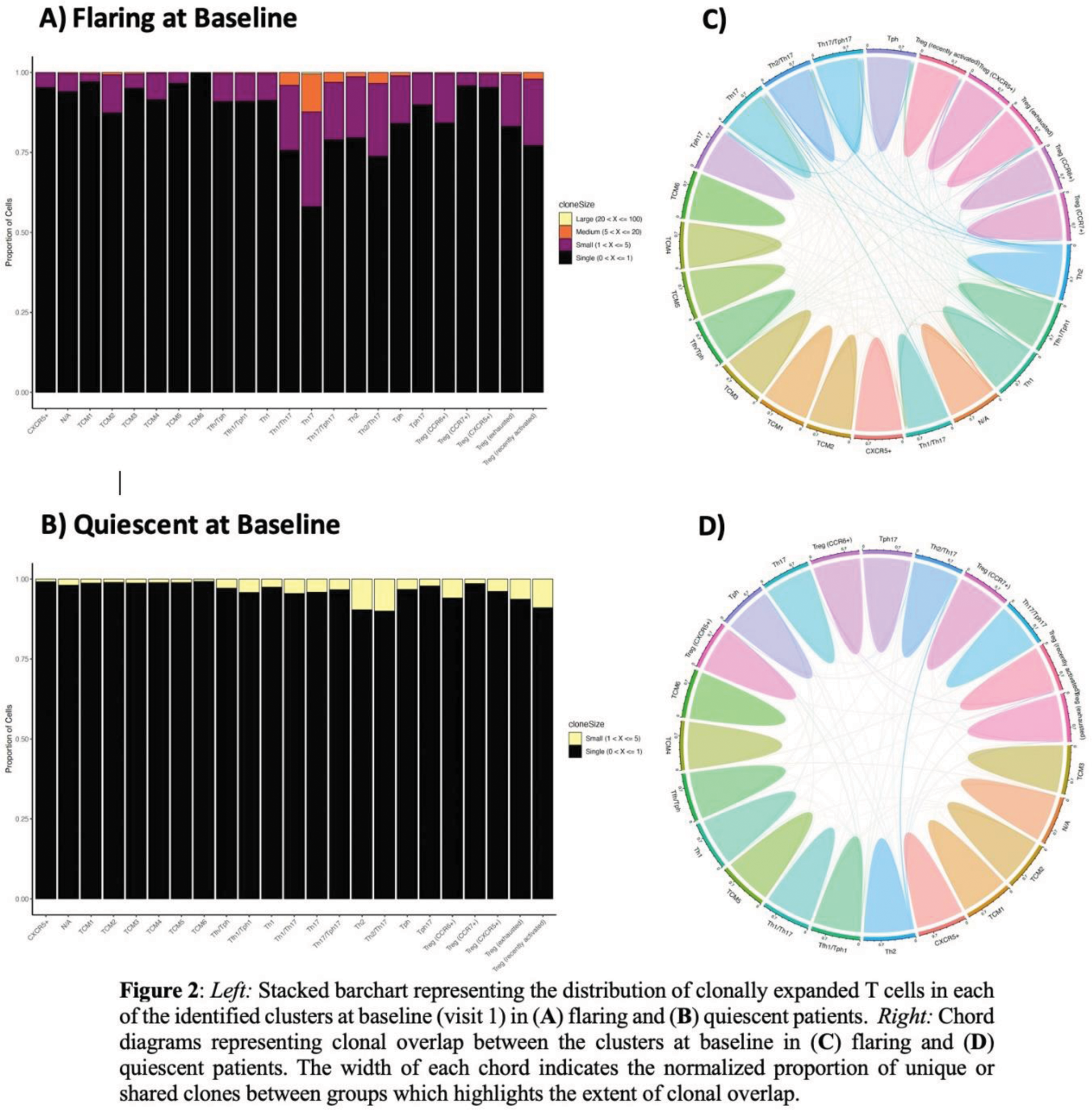
Conclusion: Although flaring patients demonstrate expansion of relatively few T cell subsets (Tfh, Tph, Th1 cells), analysis of TCR clonotypes suggests that T cells involved in the autoimmune response are found in several additional T cell subsets (Th17, Th2), indicating that there is substantial functional diversification/plasticity in the response. These findings are absent in quiescent patients, which instead show enrichment of TCMs expressing diverse TCRs. Our findings suggest that expanded populations of antigen-specific T cells can be seen in flaring patients, and that further characterization of these cells may provide insight into the specificity of the T cells that support auto-antibody production in lupus flares.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (