

Background: Psoriatic Arthritis (PsA) is a complex and heterogeneous condition, presenting considerable difficulties in its effective management. The control of disease activity is particularly challenging due to the disease’s inherent variability and multifactorial pathogenesis. Transcriptomics explores gene regulation and expression across biological stages, including treatment responses. Identifying transcriptomic biomarkers could predict drug responses, advancing personalised and stratified medicine.
Objectives: This study aimed to examine the molecular networks underlying treatment response in PsA patients undergoing secukinumab therapy.
Methods: Patients were recruited from the Outcomes of Treatment in Psoriatic Arthritis Study Syndicate (OUTPASS), a UK-based longitudinal observational study of PsA patients initiating biologic or small molecule DMARD therapy. In this transcriptomic study, inclusion criteria required PsA patients treated with secukinumab who had paired whole blood RNA samples available at baseline and 3 months along with a written consent form. Extreme response was defined as achieving a PsARC response at 3 months with a DAS-28 score <3.2, while non-response was characterised by the lowest or negative change in DAS-28 within the cohort. After whole blood RNA sequencing and quality control, differential expression analysis was performed using DESeq2 to identify gene expression signatures. Differentially expressed genes (DEGs) with a log2 fold change >1 and adjusted p-value <0.05 were further analysed using Ingenuity Pathway Analysis (IPA).
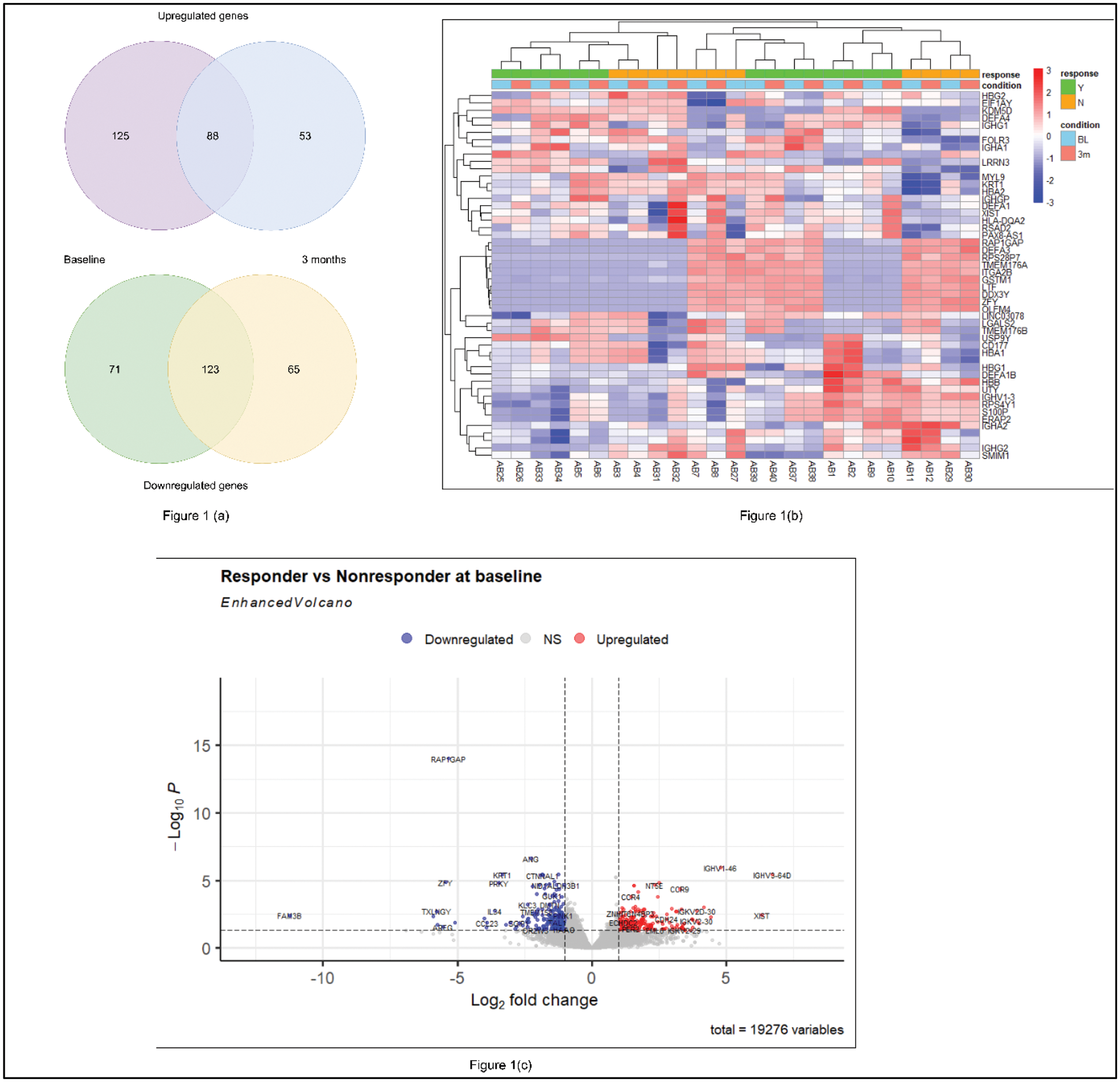
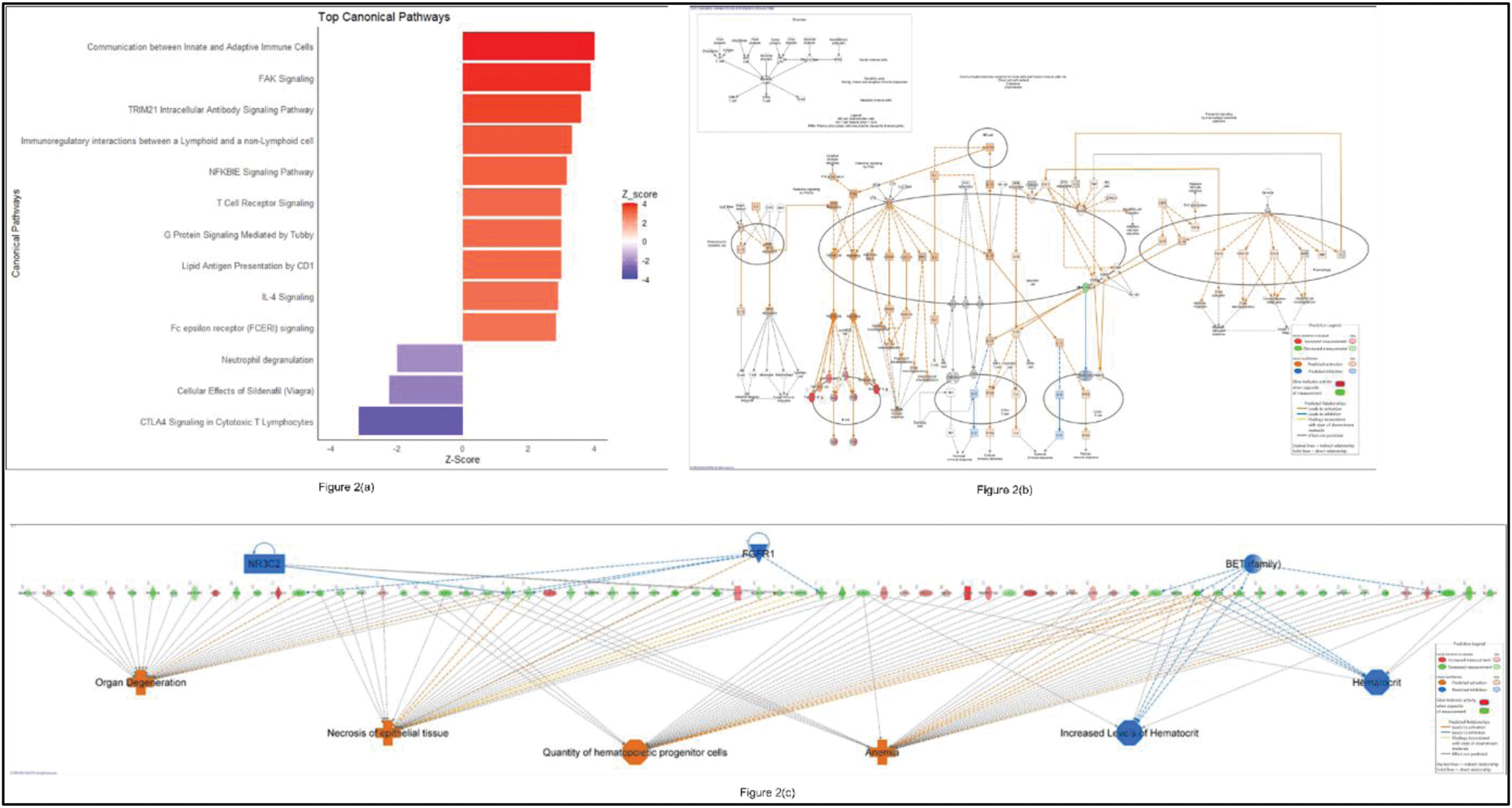
Results: Thirteen participants (responders [R], n=7; non-responders [NR], n=6) were eligible for this analysis. Transcriptomic comparison between PsA R and NR identified 407 DEGs (213 upregulated, 194 downregulated), with 325 DEGs (184 upregulated, 141 downregulated) detected at baseline and after 3 months of secukinumab therapy, respectively. Notably, 88 upregulated and 123 downregulated genes were consistent between baseline and 3 months (Figure 1(a)). Among the upregulated genes, the top five consistent DEGs were IGHV3-64D, XIST, IGHV1-46 , MDGA1 , and SCN3A . IPA of upregulated DEGs in responders at baseline highlighted the activation of pathways including communication between innate and adaptive immune cells, FAK signaling, and TRIM21 intracellular antibody signaling (z-score > 2) (Figure 2(a, b)). Additionally, gene network analysis identified key upstream regulators, such as NR3C2, FGFR1, and BET (family) (z-score >2 or < -2, p-value <0.05), suggesting their critical roles in the response to secukinumab therapy (Figure 2(c)). Time-series analysis using the likelihood ratio test revealed significant gene expression changes ( AREG, CCL23, IL34, IGHV3-64D, IGHV1-46 ) between timepoints, linked to cell-cell signalling, immune regulation, and inflammation.
Conclusion: This study characterises the transcriptomic profile of PsA patients’ response to secukinumab therapy identifying pre-treatment biomarkers predictive of response and highlighting key pathways and upstream regulators. Further external replication is needed to validate these findings.
REFERENCES: NIL.
(a) Venn Diagram (b) Heatmap of DEGs (c) Volcano plot between responders and nonresponders at baseline. The upregulated and downregulated genes are coded by red and blue points, respectively while gray points represent genes with no significant difference.
These figures illustrate molecular networks in responders at baseline. (a) Top canonical pathways. (b) Communication between innate and adaptive immune cells pathways with predicted expression changes in specific genes and proteins. (c) This figure highlights upregulated genes (displayed horizontally in the centre), their upstream regulators (above), and associated diseases and functional categories (below).
Acknowledgements: Outcomes of Treatment in Psoriatic Arthritis Study Syndicate (OUTPASS). This study has been delivered through the National Institute for Health and Care Research (NIHR) Manchester Biomedical Research Centre (BRC) (NIHR203308). The views expressed are those of the author(s) and not necessarily those of BMS, the NIHR or the Department of Health and Social Care.
Disclosure of Interests: Nahdia Afiifah Abdul Jalil: None declared , Anne Barton Research grant award from BMS, Hector Chinoy: None declared , Meghna Jani: None declared , Nisha Nair: None declared , Paul Martin: None declared , James Bluett Research grant award from Pfizer (2018) and in the last 3 years travel/conference fees from UCB (2021) and Fresenius Kabi (2022).
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (