

Background: Methotrexate (MTX), commonly used as first-line treatment for psoriatic arthritis (PsA), shows variable effectiveness, with approximately 30% of patients failing to respond within the first year due to side effects or nonresponse [1]. Limited mechanistic insight exists on how PsA patients respond to MTX. Understanding the molecular mechanisms underlying response vs nonresponse could lead to identification of clinical biomarkers, enabling improved treatment stratification and decision-making in PsA.
Objectives: To investigate the hypothesis that distinct molecular patterns drive MTX response versus nonresponse in Danish PsA patients initiating MTX as first-line therapy, using whole transcriptome single-cell RNA sequencing of peripheral blood mononuclear cells (PBMCs).
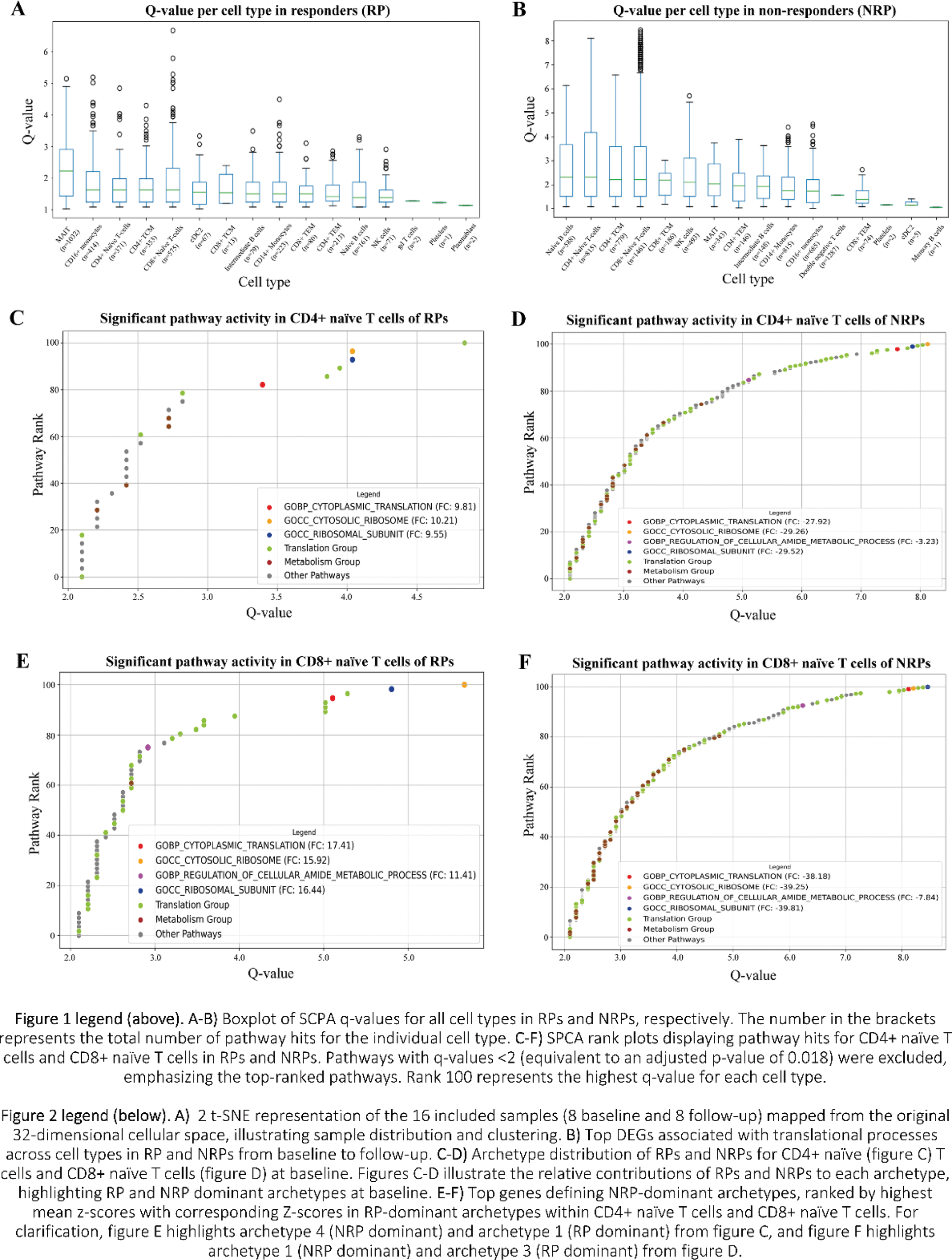
Methods: PBMCs were sampled from PsA patients (n=8) before MTX initiation and at 4 months. Samples were sorted by microwells, capturing ~8,000 cells per patient per timepoint, and sequenced at 50,000 reads per cell (paired-end, 2x150bp). Patients were classified as responders (RPs) or nonresponders (NRPs) based on Disease Activity in Psoriatic Arthritis 50 (DAPSA50). Data was analyzed using custom build annotation, Archetypal Analysis modeling, as well as Single Cell Pathway Analysis (SCPA), and differential expression (DE) analysis. Pathway hits from MSigDB Ontology Sets (C5) [2, 3] were grouped into functional categories. SCPA metrics included q-values (gene distribution changes) and fold change (FC) in pathway expression. Q-values >1.14 were considered significant (equivalent to adjusted p-values <0.05).
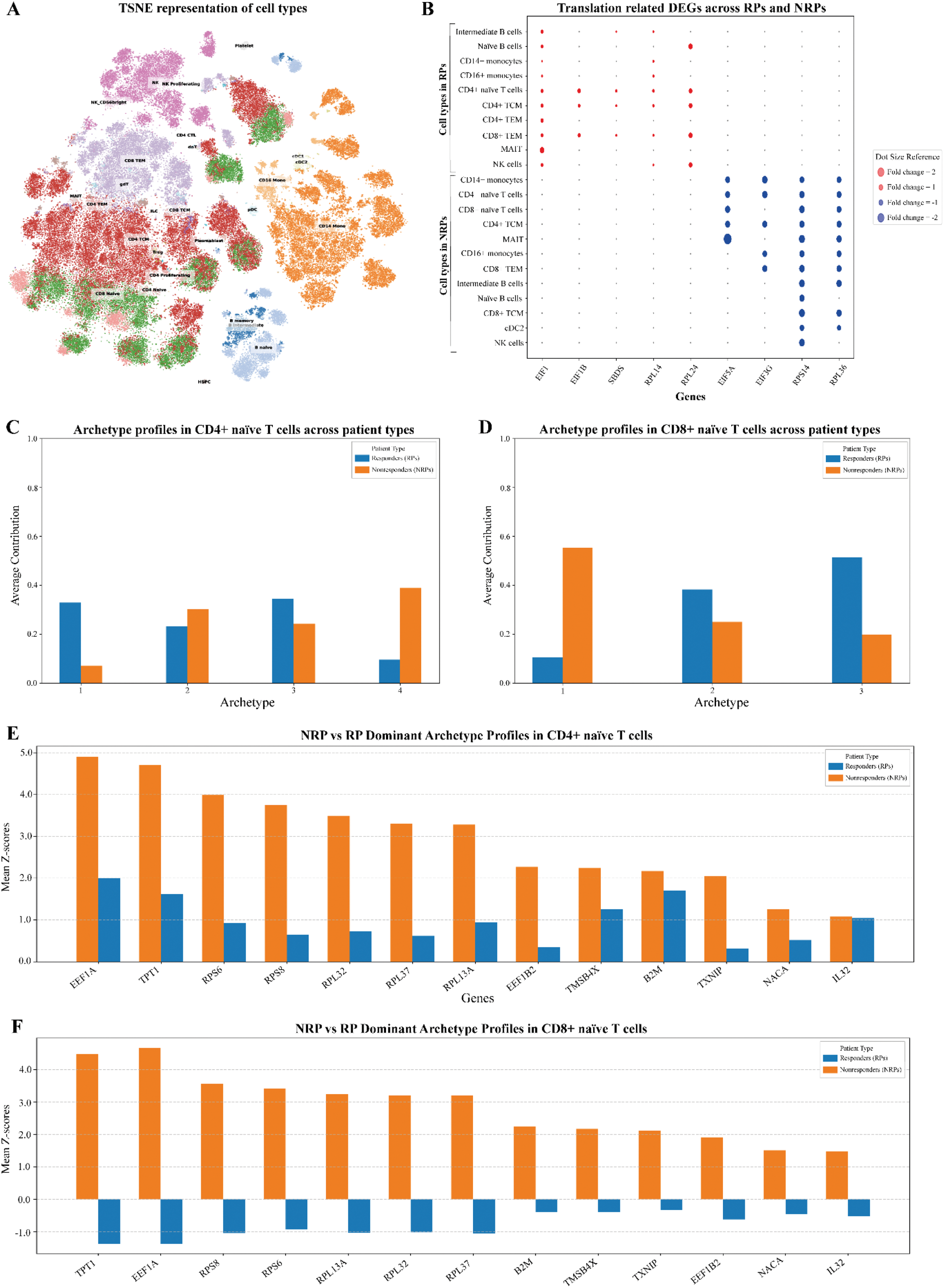
Results: Data were obtained from 5 RPs and 3 NRPs; mean DAPSA at baseline in RPs 29.7 (SD 10.6) and NRPs 25 (SD 13.7) with mean follow-up DAPSA in RPs 9.7 (SD 3.6) and NRPs 23.3 (SD 8.3). Figures 1A-B show significant pathway activity shifts toward follow-up, with CD4+ and CD8+ naïve T cells selected for further analyses due to high activity shifts. SCPA hits for mRNA translation exhibited the largest gene distribution changes (high q-values) in RPs and NRPs (Figures 1C-F). Translation pathways were upregulated in RPs (CD4+ naïve T cells [FC=9.81], CD8+ naïve T cells [FC=17.41]) but markedly downregulated in NRPs (CD4+ naïve [FC=-27.92], CD8+ naïve T cells [FC=-38.18]). Given MTX’s inhibition of purine synthesis, metabolism-related pathways were then examined. Amide metabolic processes showed the largest gene distribution changes in both groups, with downregulation in NRPs (CD4+ naïve [FC=-3.23], CD8+ naïve [FC=-7.84]) but upregulation in RPs (CD8+ naïve [FC=11.41]), though in fewer cells and with lower distribution changes compared to NRPs. DE analysis revealed upregulation of EIF1, EIF1B, SBDS, and RPL/RPS genes in RPs, while EIF5A, EIF3G, and RPL/RPS genes were markedly downregulated in NRPs (Figure 2B). EIF5A is critical for translational elongation by resolving ribosomal stalling at difficult-to-translate regions and is critically dependent on hypusination - a rare post-translational modification reliant on cellular amide metabolism. Additionally, MTX molecules undergo polyglutamation within cells, a process that could theoretically disrupt intracellular glutamate stores. As glutamate is a precursor for glutamine, a key nitrogen donor in amide metabolism, this potential disruption may contribute to amide metabolic dysregulation observed in NRPs; however, this connection requires further investigation. Archetype analysis of CD4+ and CD8+ naïve T cells at baseline (Figure 2C-D) identified NRP-dominant archetypes characterized by high expression of top genes (mean z-scores) including TPT1, RPS/RPL, B2M, TXNIP, TMSB4X, EEF1B2, NACA, and IL32 (Figure 2E). These genes play key roles in translation, oxidative stress management, and activation. Archetypes are defined as arche-cell-types present in the dataset that combinatorically describe the entire population of cells. Notably, the NRP archetype profile was consistent across CD4+ and CD8+ naïve T cells, with several genes showing markedly lower z-scores in RP-dominant archetypes, except for IL32 in CD4+ naïve T cells. The similarity across cell types highlights the potential systemic role of these archetypes in determining MTX response.
Conclusion: Our findings suggest that MTX nonresponse is influenced by a cytoplasmic ‘tipping point’ in translational control, driven by baseline cellular phenotypes that are predisposed to impaired protein synthesis under stress. Archetypes dominated by NRPs exhibited high basal activity of translational and oxidative stress-related genes, which may reflect a state of oxidative and translational pre-activation. This pre-stressed state could render NRPs less adaptable to MTX-induced challenges. Furthermore, the potential disruption of cellular glutamate stores by MTX polyglutamation could exacerbate this metabolic stress, impairing amide metabolism and subsequent hypusination of EIF5A. The concurrent downregulation of EIF5A and EIF3G in NRPs, both essential for the translation of transcripts involved in apoptosis and proliferation, could suggest Integrated Stress Response activation as a key driver of targeted translational remodelling. This response, reflecting a cellular attempt to conserve resources under stress, may impair the ability of NRPs to effectively resolve inflammation. These findings highlight the need for further validation to clarify the exact interplay between translational regulation, amide metabolism, and MTX response.
REFERENCES: [1] E. Feist, et al. J. Clin. Med. 2024;vol 13:7540, 2024.
[2] Subramanian, et al. PNAS, 2005.
[3] Liberzon, et al. Bioinformatics, 2011.
Acknowledgements: We wish to acknowledge the patients and Henning Bliddal, Director of the Parker Institute, for their contribution to the study and the parties providing funding to complete the study, including Eli Lilly (I1F-NS-O004), The Danish Rheumatism Association (R188-A6597), and the Oak Foundation through an unrestricted core grant to the Parker Institute (OCAY-18-774-OFIL).
Disclosure of Interests: Magnus Blom Petersen Research student at LEO Pharma, Oscar Møberg: None declared , Melanie Randahl Nielsen: None declared , Wenning Zheng Funding from the LEO Foundation, Albert Duvetorp Speaker fees from educational events for Abbvie, Janssen, Boehringer Ingelheim, Advisory board Eli Lilly, Almirall. Expert in development of educational material for UCB and Abbvie, Abbvie has funded research grants/donations, the Novo Nordisk Foundation funds the BRIDGE translational excellence program that A Duvetorp is part of, Jens Gerwien Shareholder at Eli Lilly, Employed at Eli Lilly, Leon Eyrich Jessen: None declared , Lars Erik Kristensen Employee at LEO Pharma, Marie Skougaard Speaker fees from Janssen Cilag, Research grants from Eli Lilly, UCB and Pfizer.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (