

Background: Up to thirty percent of people with psoriasis will develop psoriatic arthritis (PsA). There are often significant delays in the diagnosis of people with PsA. Detection of people with psoriasis who are at increased risk for developing PsA is essential to aid early diagnosis and treatment and prevent irreversible joint damage and functional disability. There are several limitations of the clinical prediction models that have been published to date to identify people with psoriasis at risk of developing PsA: none of them have been externally validated and most of them are based on studies with modest sample sizes.
To develop and internally validate a multivariable prediction model to predict the risk of developing PsA in adults newly diagnosed with psoriasis in primary care.
To externally validate the above multivariable prediction model in different populations.
Methods: A retrospective observational cohort study using primary care electronic health record (EHR) data was performed using available data from 2000-2024. The exposure cohort were adults with an incident diagnosis of psoriasis, who had been in the database for at least one year prior. The outcome was a diagnosis of PsA. The time at risk for the prediction model was five years. The Clinical Practice Research Datalink (CPRD) GOLD (UK) was used for the development and internal validation of the model; a 90:10 train/ test split by person was used for this. The proportion of the outcome was maintained when splitting by train and test datasets. The Health Improvement Network (THIN) databases from the UK, France, Spain, Italy, Romania, Belgium were used for the external validation of the model. All these EHR databases have been mapped to the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM), enabling federated analysis across the databases. A data-driven approach was used to identify the candidate predictors. All clinical variables available in primary care databases were used including demographics, co-morbidities, visit occurrence to the GP, and laboratory measurements. Extreme Gradient Boosting (XGBoost), a supervised machine learning model, was used to build the model. Model evaluation involved the area under the receiver operating characteristic (AUROC) and its 95% CI in both train and test cohorts, discrimination and calibration graphs.
Results: Table 1 summarises the results from the model development in CPRD GOLD and the external validation results in The Health Improvement Network (THIN) databases including the numbers of patients with psoriasis and PsA in each of the databases and the AUROC and its 95% CI. Figure 1 summarises the graphs for discrimination and calibration for the internal validation in CPRD GOLD. The discrimination graphs in the train and test datasets in CPRD GOLD showed that the model performed above the threshold (black dotted line). The calibration graph for the train dataset in CPRD GOLD demonstrated good calibration (the model followed the expected red dotted line). However, this was not the case for the test dataset in CPRD GOLD which showed poor calibration: the model underpredicted the outcome. This was also the case for the calibration in all the THIN databases where the model was externally validated: it showed poor calibration. (Figure 1) The predictors with the highest discriminatory power in this model were age and visit occurrence to the GP in the one year/ six months/ 1 month prior to their psoriasis diagnosis.
Results from the model development and external validation
| Database name | Number of psoriasis patients | Number of psoriatic arthritis outcomes (proportion from total patients) | Area under the receiver operating characteristic (AUROC) | 95% lower Confidence Interval | 95% upper Confidence Interval |
|---|---|---|---|---|---|
| Model development in Clinical Practice Research Datalink (CPRD) GOLD | |||||
| CPRD Train | 150,817 | 2,651 (1.76%) | 0.78 | 0.78 | 0.79 |
| CPRD Test | 16,758 | 295 (1.76%) | 0.68 | 0.65 | 0.71 |
| External validation in The Health Improvement Network (THIN) databases | |||||
| THIN UK | 235,192 | 6,506 (2.8%) | 0.63 | 0.62 | 0.64 |
| THIN Spain | 26,260 | 1,044 (4.0%) | 0.54 | 0.51 | 0.56 |
| THIN France | 138,312 | 8,958 (6.5%) | 0.48 | 0.47 | 0.49 |
| THIN Italy | 6,946 | 290 (4.2%) | 0.56 | 0.51 | 0.61 |
| THIN Romania | 6,620 | 626 (9.5%) | 0.52 | 0.49 | 0.56 |
| THIN Belgium | 20,642 | 1,008 (4.9%) | 0.50 | 0.48 | 0.53 |
discrimination and calibration graphs for the internal validation in CPRD GOLD 1
1 The black dotted line in the discrimination graphs represents an area under the receiver operating characteristic (AUROC) of 0.5, which is the same as the performance of a random model. The red dotted line in the calibration graphs represents the perfect calibration (i.e., the values between observed and predicted probability would been the same): a good model will have a line close to the perfect calibration.
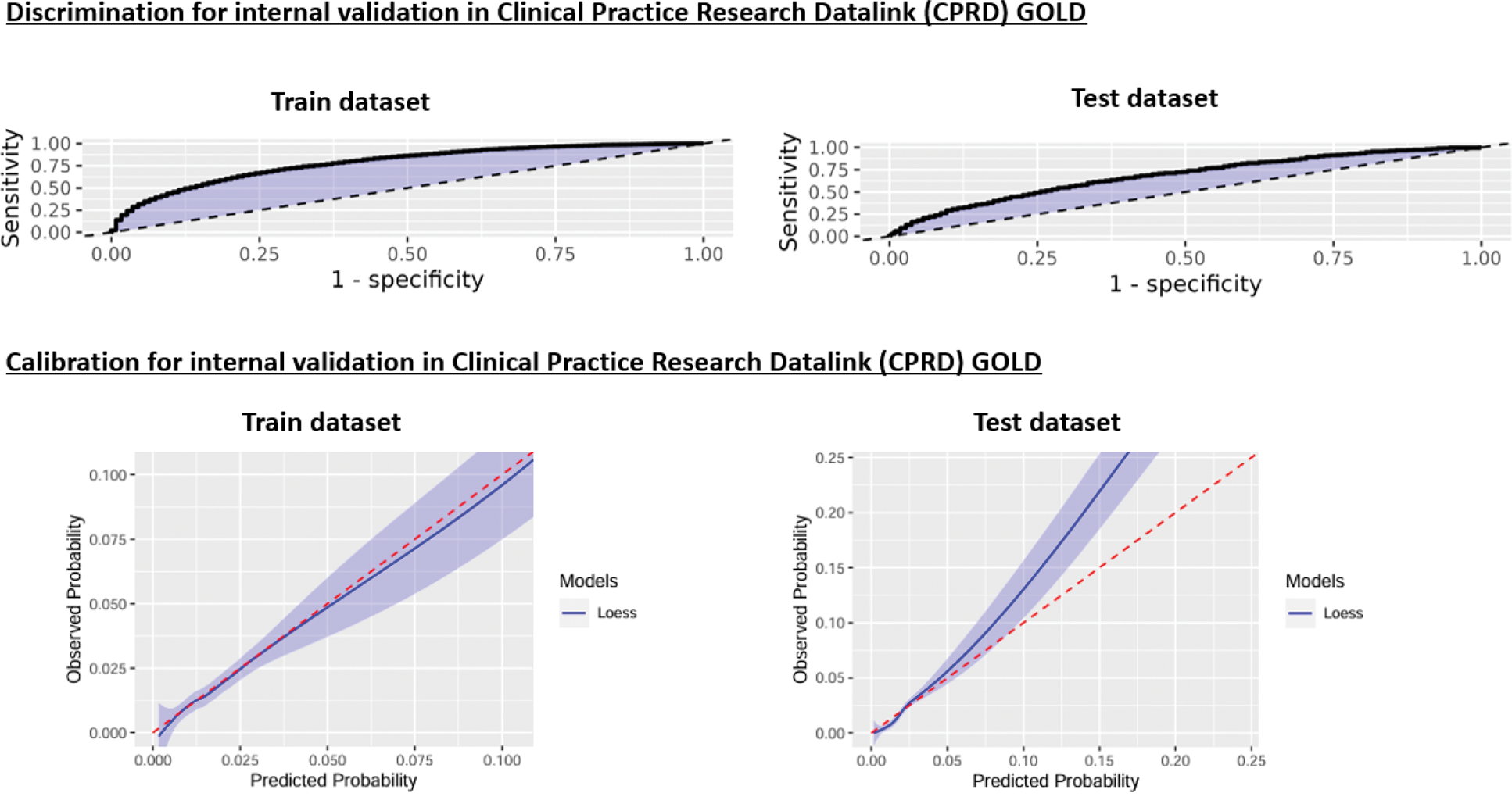
Conclusion: To the best of our knowledge, this is the first clinical prediction model for predicting PsA developed using clinical markers available in primary care that has been externally validated. The model performs well in the dataset it was developed in (with a similar performance to other models that have been published). However, we found that the model did not perform well when externally validated. It could be that further granular data (e.g., nail psoriasis and Psoriasis Area and Severity Index score) that are not available in primary care data and other markers (e.g. genetic) are needed to predict PsA. The Health initiatives in Psoriasis and PsOriatic arthritis ConsoRTium European States (HIPPOCRATES) consortium, funded by the EU, is aiming to address this through the HIPPOCRATES Prospective Observational Study (HPOS), which is a prospective cohort study running across Europe, collecting granular data on patients at the inception of their psoriasis diagnosis with the intention to identify those at risk of developing PsA.
REFERENCES: NIL.
Acknowledgements: AV is funded by an NIHR Doctoral Research Fellowship (Award ID: NIHR302335). This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement No 101007757. The JU receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA.
Disclosure of Interests: Arani Vivekanantham: None declared. Marta Pineda-Moncusí: None declared. Edward Burn: None declared. Sara Khalid: None declared. Daniel Prieto-Alhambra DPA has provided consultancy services, with fees paid to the University, for UCB Biopharma. DPA’s department has received grants from Amgen, Chiesi-Taylor, Gilead, Lilly, Janssen, Novartis, and UCB Biopharma. Janssen has funded or supported training programmes organised by DPA’s department. DPA is a member of the Board of the EHDEN Foundation and has received grants from the European Medicines Agency and the Innovative Medicines Initiative. Laura C. Coates LCC has been paid as a speaker for AbbVie, Amgen, Biogen, Celgene, Eli Lilly, Galapagos, Gilead, GSK, Janssen, Medac, Novartis, Pfizer and UCB. LCC has worked as a paid consultant for AbbVie, Amgen, Bristol Myers Squibb, Celgene, Eli Lilly, Gilead, Galapagos, Janssen, Moonlake, Novartis, Pfizer, Takeda and UCB, LCC has received grants/research support from AbbVie, Amgen, Celgene, Eli Lilly, Janssen, Novartis, Pfizer and UCB. LCC is supported by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre (BRC).
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (