

Background: Treatment response in lupus nephritis (LN) remain inadequately low, highlighting the need for better understanding of LN pathogenesis to improve management. Single-cell transcriptomic studies are providing an unprecedented catalog of cell states in LN, yet their spatial organization is not well understood. Since structure underlies function, we aim to map the spatial organization of immune cells in LN.
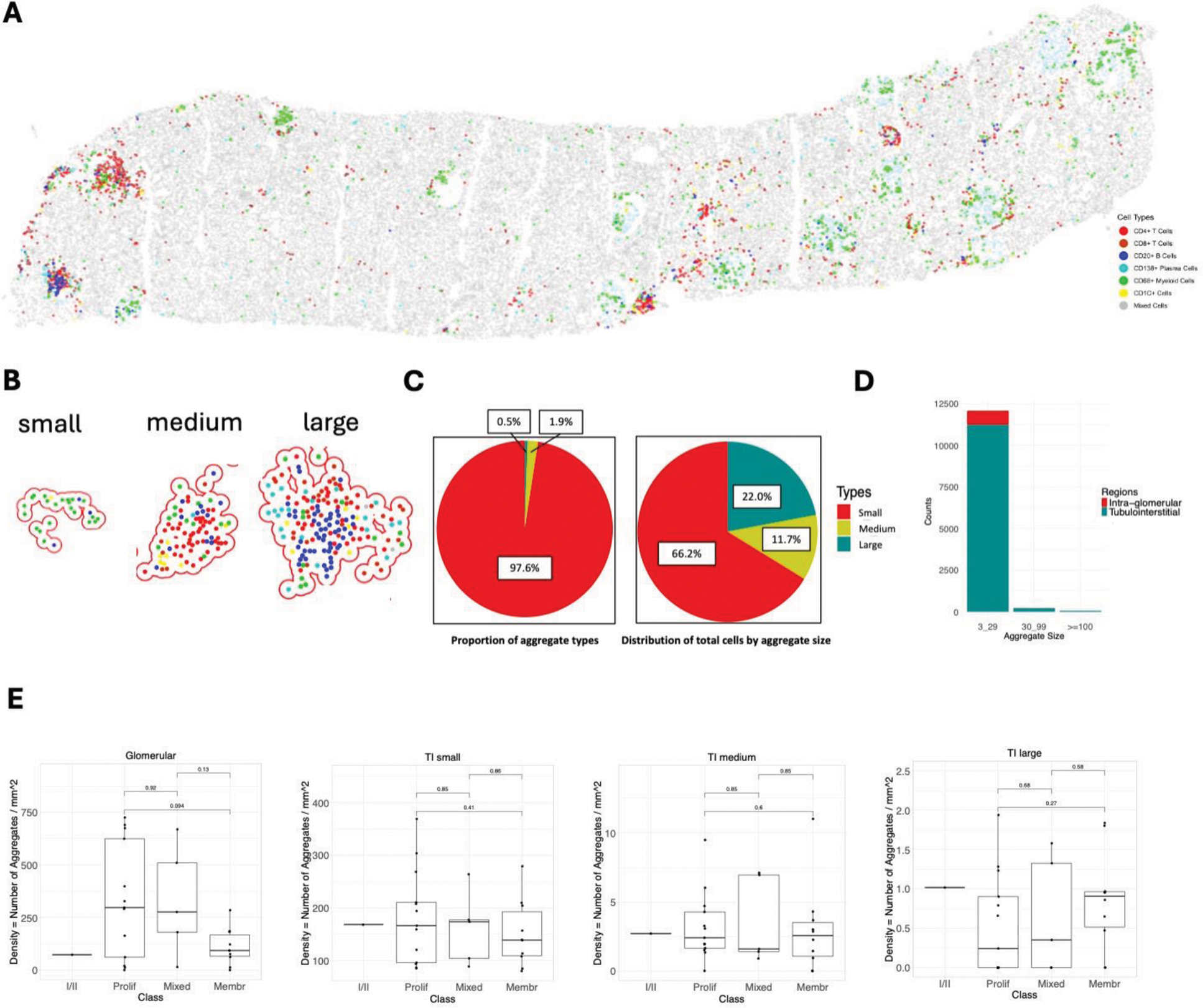
Objectives: We developed a serial immunohistochemistry (sIHC) workflow (18-plex), followed by imaging and destaining cycles. Image processing was performed using HALO (Indica Labs), including AI-assisted tissue classification. PCA was used for dimensional reduction, and KNN and SNN algorithms were applied to identify immune cell types based on their markers’ fluorescent intensity. Immune cell aggregates in the tissues were defined using DBSCAN as a minimum of 3 cells within a radius (epsilon) of 50 μm to infer interactions between cells. Aggregate sizes were categorized into small (3-29 cells), medium (30-99 cells), and large (>100 cells) based on the frequency distribution (Figure 1A-B). The proportion of immune cell types in each aggregate was used for K-means clustering to determine aggregate subtypes. Clinical features were correlated with each aggregate subtype using Pearson’s correlation coefficient (Figure 2).
Methods: We developed a serial immunohistochemistry (sIHC) workflow (18-plex), followed by imaging and destaining cycles. Image processing was performed using HALO (Indica Labs), including AI-assisted tissue classification. PCA was used for dimensional reduction, and KNN and SNN algorithms were applied to identify immune cell types based on their markers’ fluorescent intensity. Immune cell aggregates in the tissues were defined using DBSCAN as a minimum of 3 cells within a radius (epsilon) of 50 μm to infer interactions between cells. Aggregate sizes were categorized into small (3-29 cells), medium (30-99 cells), and large (>100 cells) based on the frequency distribution (Figure 1A-B). The proportion of immune cell types in each aggregate was used for K-means clustering to determine aggregate subtypes. Clinical features were correlated with each aggregate subtype using Pearson’s correlation coefficient (Figure 2).
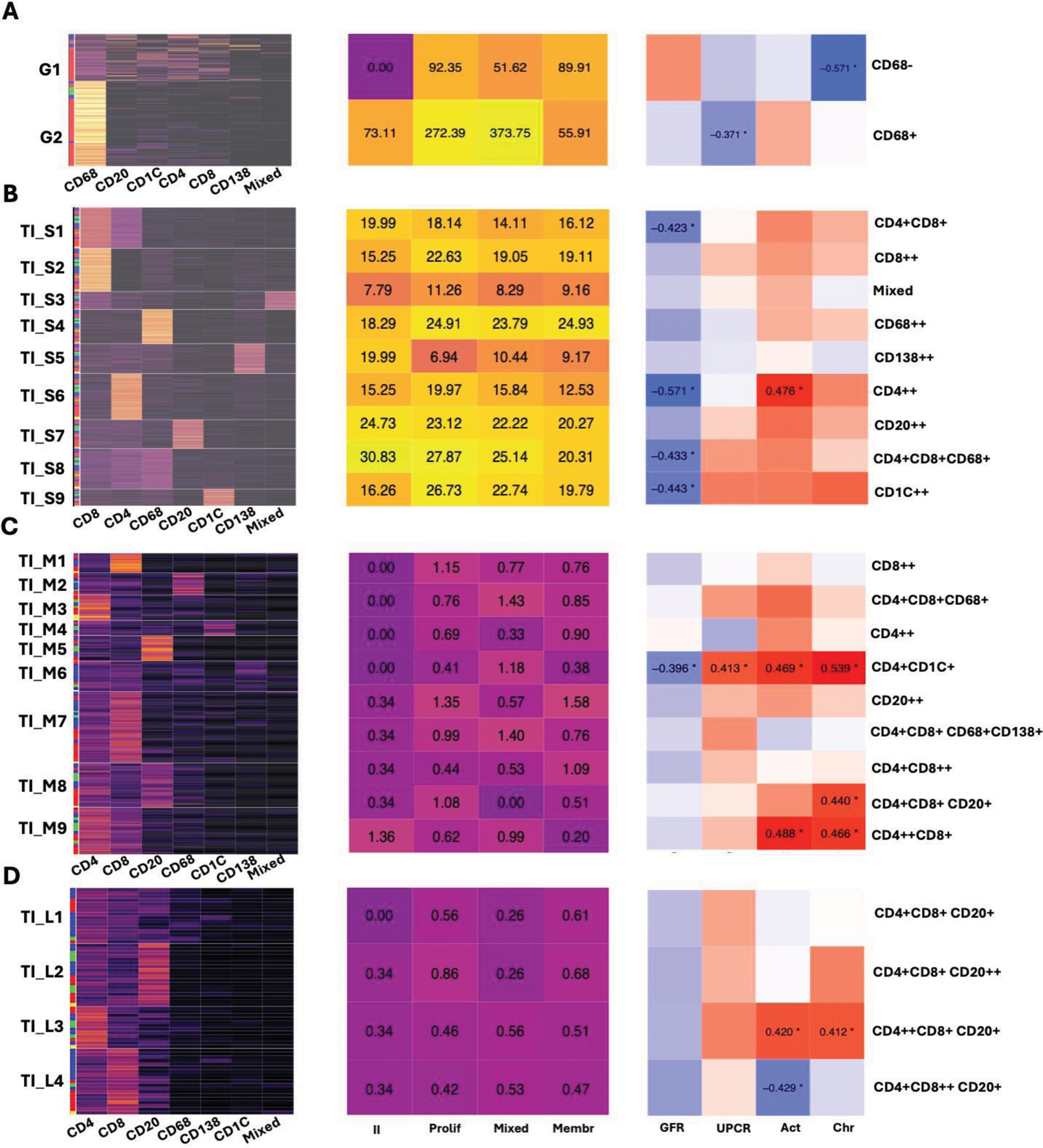
Results: In this analysis, we included 29 kidney biopsies of LN resulting in 1,913,845 cells (182,783 immune cells). We identified 12,371 cellular aggregates. Most (97%) aggregates were small (<30 cells) (Figure 1C-D); however, medium and large aggregates included 33.7% of immune cells. Glomerular aggregates were numerically increased in proliferative and mixed classes (Figure 1E). These were small and primarily composed of CD68+ myeloid cells (Figure 2A). Glomerular aggregates rich in CD68+ cells negatively correlated with UPCR, while aggregates rich in lymphocytes negatively correlated with chronicity (Figure 2A). In contrast, tubulointerstitial (TI) aggregate density was similar across LN classes (Figure 1E) and negatively correlated with GFR. Significant heterogeneity in aggregate composition revealed >10 aggregate subtypes according to composition and size (Figure 2). Small aggregates tended to be restricted to 1-2 cell types each, while medium and large aggregates included mixed proportions of CD4+ T, CD8+ T, B, dendritic, myeloid, and plasma cells, suggesting germinal center-like structures (Figure 2). Distinct TI aggregate subtypes associated with specific clinical and pathological features (Figure 2B-C).
Conclusion: We describe the heterogeneity in glomerular and TI immune cell structures in LN, offering insights into LN pathological processes and potential cellular interactions based on proximity. TI inflammation appears similar in membranous and proliferative LN, yet specific immune structures are linked to distinct clinical and pathological features.
Demographics of intrarenal immune cell aggregates. (A) Digitalized biopsy showing an example of the distribution of immune cells in LN. (B) Examples of intrarenal immune cell aggregates of different sizes. (C) Distribution of aggregates by size and by total cells. (D) Distribution of aggregates by size and region. (E) Density of aggregate types according to size and class.
Correlation between aggregate subtypes and clinical features. Heatmaps on the left displaying the subtypes of immune cell aggregates. The proportion of immune cell types in each aggregate was used for K-means clustering to determine aggregate subtypes. The column annotation rows of the heatmaps represent LN classes, red: proliferative; green: mixed, blue: pure membranous; yellow: ISN class II. Heatmaps in the middle show the average density of different aggregates, measured as the average number of aggregates per mm². Heatmaps on the right showing the correlation matrices between the aggregate subtypes and clinical features. Clinical features, including GFR, urinary protein-creatinine ratio (UPCR), and NIH activity and chronicity indices, were correlated with each aggregate subtype using Pearson’s correlation coefficient. Correlation coefficients with statistical significance are shown and marked with asterisks. (A) Glomerular small aggregate (B) Tubulointerstitial small aggregate (C) Tubulointerstitial medium aggregate (D) Tubulointerstitial large aggregate. Act: NIH activity index; Chr: NIH chronicity index
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: CHEN-YU LEE: None declared. Matthew Caleb Marlin: None declared. Xiaoping Yang: None declared. Vasileios Morkotinis: None declared. Alessandra Ida Celia: None declared. Jill Buyon Artiva Biotherapeutics, Bristol-Myers Squibb (BMS), Equillium, GlaxoSmithKlein (GSK), Otsuka Pharmaceuticals, Related Sciences, Bristol-Myers Squibb (BMS), GlaxoSmithKlein (GSK), Related Sciences, Richard A. Furie AstraZeneca, AstraZeneca, AstraZeneca, Diane L. Kamen Alpine Immune Sciences, Bristol Myers Squibb (BMS), Jeffrey Hodgin: None declared. Chaim Putterman Equillium, Progentec, Jennifer Barnas: None declared. Kenneth C. Kalunian: None declared. Peter Izmirly Hansoh Bio, Betty Diamond alpine, adicet, alpine, dsmb, atara, DBV, icell, sail, Anne Davidson EMD Serono, the Accelerating Medicines Partnership in RA/SLE: None declared. Judith A. James GlaxoSmithKlein (GSK), Progentec Diagnostics, Inc. Michelle Petri CTI, CVS Health, Emergent Biosolutions, IQVIA, Merck/EMD Serono, Worldwide Clinical Trials, Amgen, AnaptysBio, Annexon Bio, AstraZeneca, Atara Biosciences, Autolus, Avoro Ventures, Biocryst, Boxer Capital, Cabaletto Bio, Caribou Biosciences, Eli Lilly, Ermiium, Escient Pharmaceuticals, Exo Therapeutics, Gentibio, GlaxoSmithKlein (GSK), iCell Gene Therapeutics, Innovaderm Research, Kira Pharmaceuticals, Nexstone Immunology, Nimbus Lakshmi, Novartis, PPD Development, Precision Biosciences, Proviant, Regeneron Pharmaceuticals, Sanofi, Seismic Therapeutic, Senti Bioscienes, Sinomab Biosciences, Takeda, Tenet Medicines Inc, TG Therapeutics, UCB, Vertex Pharmaceuticals, Zydus, AstraZeneca, Aurinia, Eli Lilly, Exagen, GlaxoSmithKlein (GSK), Janssen, Arthros-FocusMedEd, Joel M Guthridge AstraZeneca, Bristol-Myers Squibb (BMS), Avi Rosenberg: None declared. Andrea Fava Annexionbio, Arctiva, AstraZeneca, UCB, Exagen, Novartis.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (