

Background: Systemic lupus erythematosus (SLE) is a complex autoimmune disease that affects multiple organ systems, complicating clinical trial design. Identifying biomarkers associated with distinct clinical features may enable more targeted drug development. The transcriptional dysregulation of key immune pathways in whole blood of SLE patients is well established [1] suggesting the ability to identify patient molecular subtypes based on blood transcriptome [2]. However, the characterization of such subtypes remains limited due to the use of small patient cohorts, few clinical features, and limited data modalities. This gap limits our understanding of the SLE molecular subtypes and our ability to use them to inform treatment strategies and clinical trial design.
Objectives: This study aims to further characterize SLE molecular subtypes using clinical trials (ILLUMINATE) [3] and a large observational research cohort (Genuity Science, Ireland), leveraging a comprehensive dataset that includes clinical, transcriptomic, proteomic, and genomic information.
Methods: Transcriptomic network analysis of whole blood identified numerous co-expressed gene modules associated with SLE [1]. These gene modules have been categorized into nine distinct signatures [4]. For this analysis, a representative module for each signature was selected, and their enrichment scores from gene expression datasets were computed using Gene Set Variation Analysis (GSVA) for 1,760 and 812 patients from the ILLUMINATE studies (whole blood) and Genuity Science cohort (PBMC), respectively. Based on module GSVA scores, ILLUMINATE patients were clustered using a consensus clustering approach to identify SLE molecular subtypes. A machine learning model was developed based on ILLUMINATE patients’ data and used to map 812 patients from the Genuity Science cohort into these subtypes. Clinical characterization of these subtypes utilized data from both the ILLUMINATE and Genuity Science cohorts. Serum proteomic profiles of the Genuity Science cohort were obtained using the Olink Explore platform. A genetic risk score was computed using PRS-CSx [5] based on the data from Bentham et al [6] for each patient in the Genuity cohort and examined across clusters.
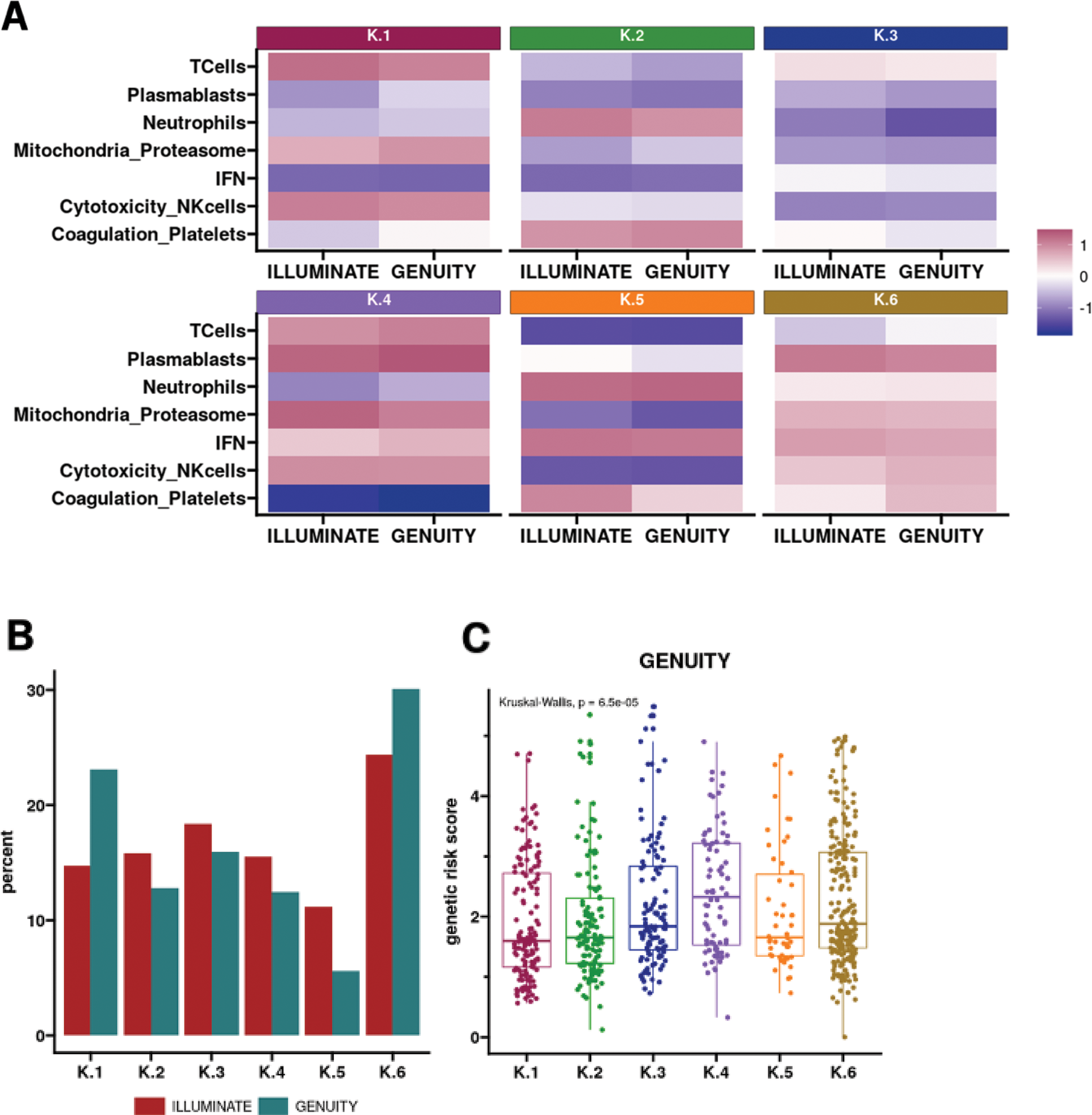
Results: Six distinct clusters were identified in the ILLUMINATE population, and these clusters mapped well to the Genuity Science cohort, with at least half of patients having at least 50% probability of being classified to their respective cluster, with all patients having at least 25% probability, compared to a 16% chance if classified randomly. The transcriptomic profiles across the 6 clusters of patients were consistent between the two cohorts (Figure 1A). The frequency of patients across the clusters varied in the Genuity Science compared to ILLUMINATE cohort (Figure 1B), reflecting overall differences between observational and clinical trial patient populations. These clusters, in both cohorts, exhibited similar differences in SLEDAI disease activity with Clusters 1, 2, and 3 exhibiting lower scores compared to Clusters 4, 5, and 6 (P<0.001). Additionally, in the Genuity Science cohort, Cluster 4 had the highest genetic risk score (Figure 1C). Strong correlations were observed with serological activity, measured by anti-dsDNA positivity (e.g., 35% in Cluster 1 vs. 71% in Cluster 6 in ILLUMINATE; 27% vs. 46% in Genuity Science cohort) and low complement levels (e.g., 24% vs. 57% in ILLUMINATE; 28% vs. 40% in Genuity Science cohort). The Genuity Science cohort revealed distinct distributions of other SLE-associated autoantibodies across clusters, namely, anti-Ro and anti-Sm. For anti-Ro, Cluster 2 had the lowest prevalence at 12%, while Cluster 3 had the highest at 43%. Similarly, for anti-Sm, Cluster 2 exhibited the lowest prevalence at 6%, with Cluster 4 showing the highest at 21%. Olink profiles exhibited unique protein abundance patterns between the clusters and consistent with whole blood transcriptomic data: IFN pathway proteins are highest in Clusters 4, 5, and 6 but lowest in Clusters 1 and 2; B cell soluble receptors and LAG3 are higher in Cluster 4; and PADI4 and OSM are higher in Cluster 2.
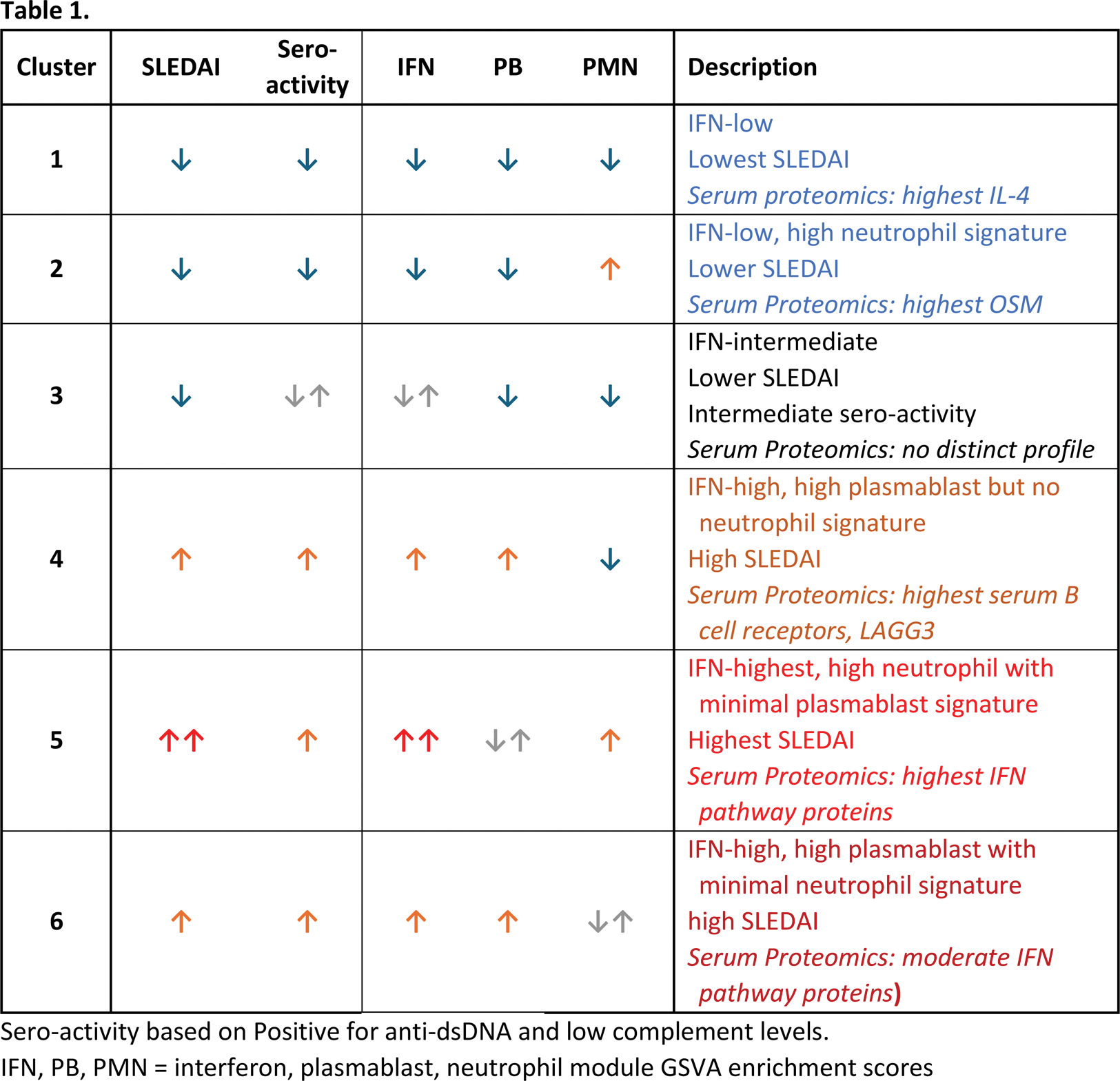
Conclusion: Using two large SLE cohorts, we identified and confirmed six molecular subtypes based on whole blood transcriptomes and conducted an in-depth analysis of their molecular, clinical, and proteomic characteristics (Table 1). Interestingly, the clusters exhibited significant differences in disease severity, genetic risk scores, serological activity, and proteomic signatures. These findings provide valuable insights for patient stratification in clinical trials and may enable the development of targeted treatment strategies based on predicted responses to immune-modulating therapies.
REFERENCES: [1] Chaussabel et al Immunity. 2008; 29(1):150-64.
[2] Hubbard et al Genome Med 2023, 84
[3] Hoffman RW, et al. Arthritis Rheumatol. 2017. 69(3):643-654.
[4] Toro-Domínguez D, et al. Brief Bioinform. 2022. 23(5):bbac332
[5] Ruan Y, et al. Nature Genetics, 54:573-580, 2022
[6] Bentham et al Nat Genet. 2015; 47(12): 1457–1464
Acknowledgements: Data from the Genuity Science research project was used in this study. We would like to express our gratitude to Genuity Science, the patients who contributed samples and data, and to the Principal Investigators, their research teams and Institutions who participated in the Genuity Science research project.
Disclosure of Interests: Loqmane Seridi Janssen Research & Development, LLC, a Johnson & Johnson Company, Janssen Research & Development, LLC, a Johnson & Johnson Company, Eugene Myshkin Janssen Research & Development, LLC, a Johnson & Johnson Company, Janssen Research & Development, LLC, a Johnson & Johnson Company, Erika Noss Janssen Research & Development, LLC, a Johnson & Johnson Company, Janssen Research & Development, LLC, a Johnson & Johnson Company, Dawn Waterworth Janssen Research & Development, LLC, a Johnson & Johnson Company, Janssen Research & Development, LLC, a Johnson & Johnson Company, Matthew J Loza Janssen Research & Development, LLC, a Johnson & Johnson Company, Janssen Research & Development, LLC, a Johnson & Johnson Company, Sheng Gao Janssen Research & Development, LLC, a Johnson & Johnson Company, Janssen Research & Development, LLC, a Johnson & Johnson Company.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (