

Background: Osteoarthritis (OA) is the most prevalent chronic joint condition, marked by structural damage to cartilage and bones, often leading to pain and disability. However, there is no clear link between the extent of structural joint damage and the level of pain experienced; up to 50% of individuals without joint symptoms may still exhibit radiographic signs of OA. Currently, three distinct subgroups of osteoarthritis (OA) patients are proposed: Pain-Progressor OA, Structural-Progressor OA, and Pain+Structural Progressor OA. The limited success of therapeutic interventions in managing OA pain may be due to the difficulty in identifying patients at high risk for both pain and structural progression; however, distinguishing between rapid structural and non-rapid structural progressors could enhance the management of OA pain. Machine learning generates predictive models that are not only accurate but also human-readable and easily automated. Recent advancements in the analysis of biomedical data, through rigorous statistical methods, allow for the extraction of complex multivariate patterns from the rule-based models generated. In OA, clinical and imaging data, as well as molecular biomarkers, have allowed our group to define a panel of biomarker candidates (20 proteins, 8 nuclear SNPs, and 3 mitochondrial haplogroups) related to OA. This can facilitate the development of knowledge extraction algorithms that identify and refine patterns linked to different subgroups within the cohort, ultimately helping to distinguish distinct patient phenotypes.
Objectives: Our objective is to develop a predictive model for the rapid structural progression of osteoarthritis by integrating various machine learning techniques and using clinical, genomic, proteomic, and epigenetic data to predict structural progression over time in well-characterized prospective patient cohorts, such as OAI and PROCOAC, while evaluating the effectiveness of this tool in both cohorts.
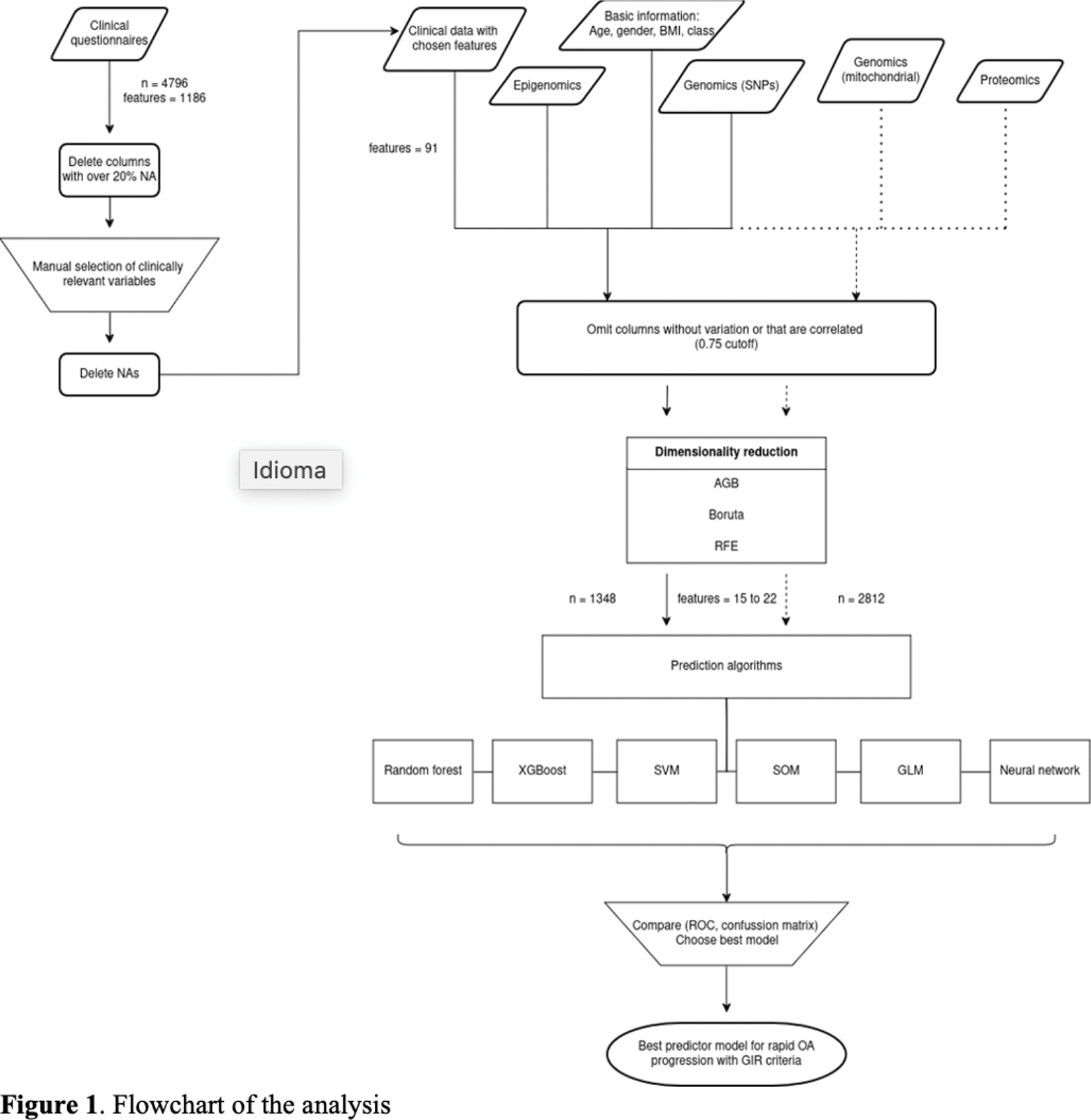
Methods: Rapid structural progression was defined according the GIR criteria, patients with an increase in the Kellgren-Lawrence (KL) score from ≤1 to ≥3, or from KL = 2 to KL = 4, or joint replacement over a follow-up period of 48 months. We used different clinical, genomic, proteomic, and epigenetic data, which were subsequently filtered using a feature selection strategy to construct the models. As part of the genomic variables, we included significant SNPs (p < 10⁻⁷) resulting from GWAS performed in the OAI database. The Polygenic Risk Score (PRS) was also defined. Six machine learning models were studied, with each database divided into 75% for training and 25% for testing to identify the best model for prediction. A flowchart is presented in Figure 1.
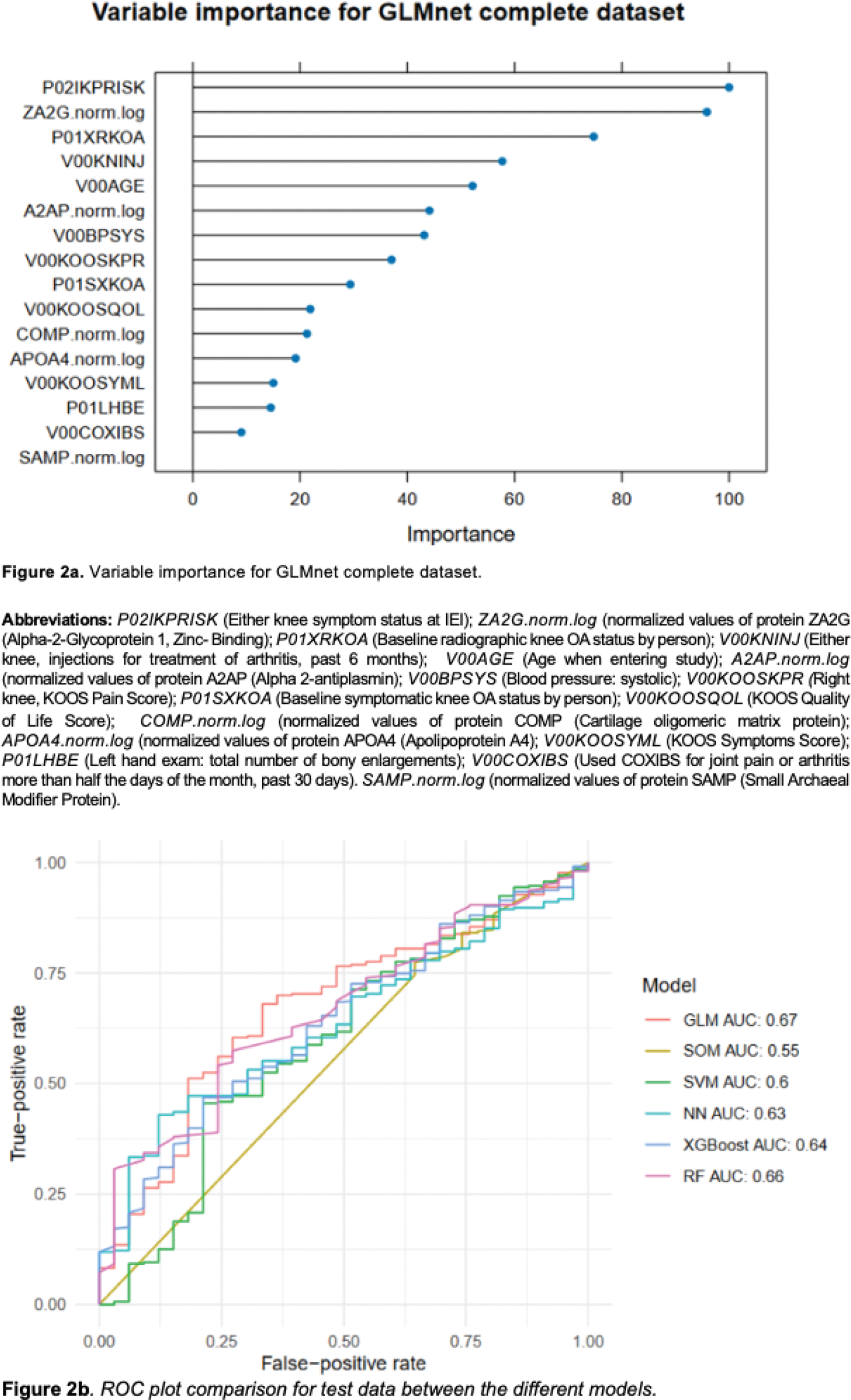
Results: GWAS analysis in the OAI showed, as the top five most significant SNPs: rs34968734 at CPNE4 gene, rs143480123 at NPCC , rs151232509 at CTD-2015G9.2 , rs117288372 at RP11-324L3.1 and rs149600528 at ZNF823 . Lassosum’s split validation method was used, splitting the original OAI cohort into two for validation and calculation of the polygenic risk score (PRS). The scores proved significant in the model based on the coefficients and predictive power of a model created with this PRS, as well as age (AUC 0.552, p < 0.001), sex (AUC 0.541, p < 0.001), and BMI (AUC 0.575, p < 0.001) of the cohort, with a global AUC of 0.694. Using Recursive Feature Elimination (RFE) and, after a preprocessing step, we reduced the number of variables from thousands to 44 with potential to provide more information about the cohort, and obtained a resulting database containing 2812 individuals, with 240 rapid progressors. Then, after applying different machine learning algorithms, we identified 16 predictive variables (Figure 2a). The best-performing algorithm during training and test was GLMnet, with an AUC of 0.760 and 0.67 respectively, as shown in Figure 2b.
Conclusion: While promising, its full potential requires validation with additional cohorts to ensure robustness. This tool could prove invaluable for the early identification of patients at risk of rapid structural progression of OA, enabling timely interventions that could significantly improve both patient outcomes and the effectiveness of early treatment strategies.
Pfizer grand ADVANCE Awards Nº Grand Agreement NPM/64122119.
REFERENCES: NIL.
Acknowledgements: Biomedical Research Institute of A Coruña (INIBIC).
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (