

Background: Systemic sclerosis (SSc) is a multi-organ autoimmune disease characterized by three main pathological hallmarks: vasculopathy, fibrosis, and autoimmunity. Despite advances in treatment and prognosis, some patients develop early organ involvement. Patient stratification is critical to identifying individuals at higher risk of organ damage, enabling earlier detection and potentially preventing its progression.
To identify distinct clinical subgroups in SSc using a clustering algorithm based on clinical and serological variables.
To compare the proteome of the identified clinical clusters to explore biomarkers associated with organ damage.
Methods: A cross-sectional observational study was conducted with 402 patients diagnosed with SSc, utilizing data from the international multicenter PRECISESADS study. Clinical and serological variables were collected. A k-means clustering algorithm was applied, integrating various clinical and serological variables. The optimal number of clusters was determined using the silhouette width method. Subsequently, a univariate analysis was performed to compare the clusters. Following the identification of clinical clusters, a random sample of 144 patients (77 from each cluster) was selected for proteomic analysis of 92 proteins associated with organ damage using OLINK technology.
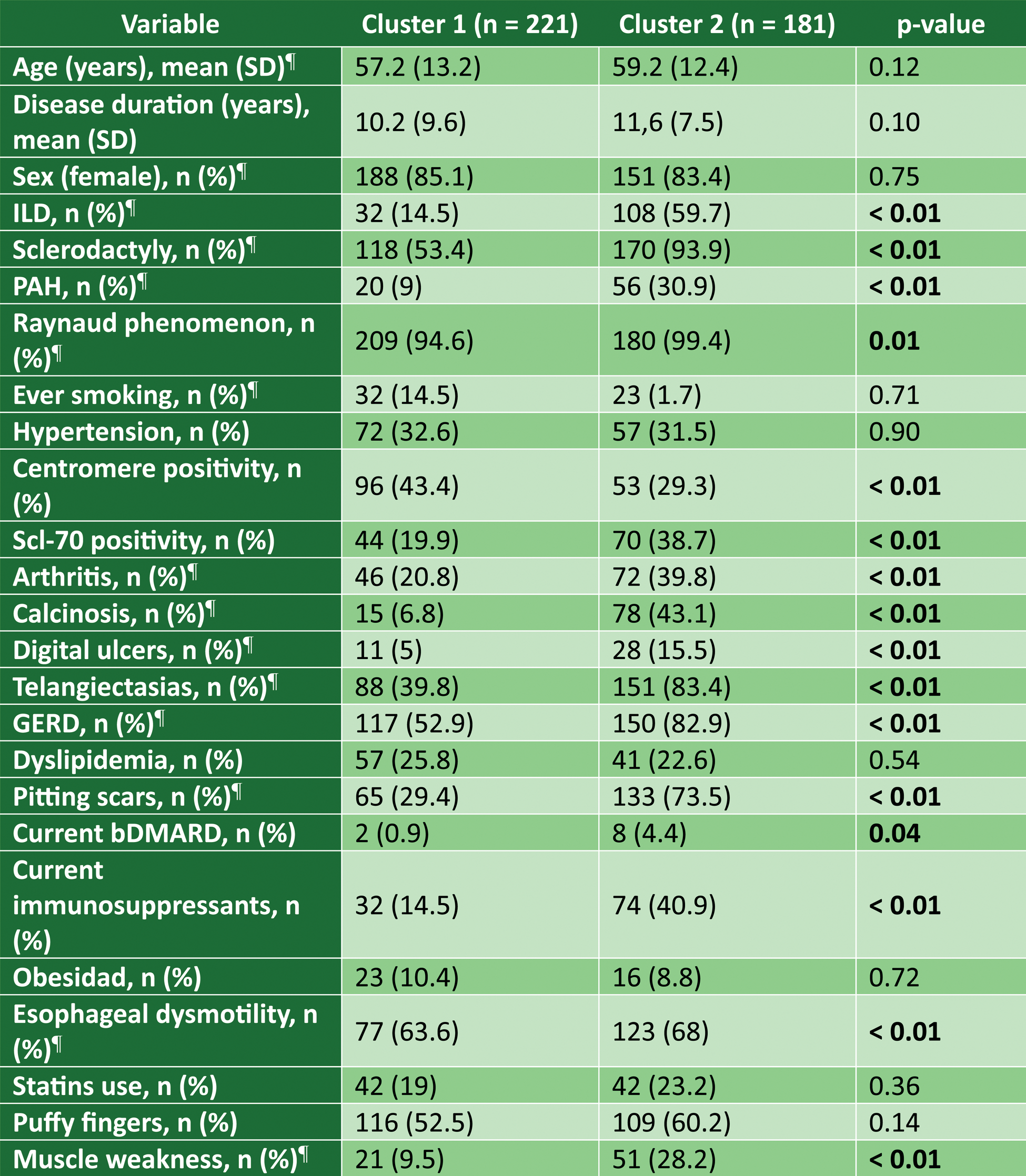
Results: Two clusters were identified (Cluster 1 = 221 (55%) and Cluster 2 = 181 (45%)). The results of the univariate analysis are shown in Table 1. Cluster 1 included a slightly younger population (57.2 vs. 59.1 years) and a shorter disease duration (10.2 vs. 11.6 years), although these differences were not statistically significant. Patients in Cluster 2 exhibited a higher prevalence of organ involvement, including ILD (14.48% vs. 59.67%), PAH (9.05% vs. 30.94%), and esophageal dysmotility (34.84% vs. 67.96%). Serologically, anticentromere antibody positivity was more frequent in Cluster 1 (43.44% vs. 29.28%), whereas anti-Scl70 positivity was more common in Cluster 2 (19.91% vs. 38.67%). Proteomic comparison between the two clusters (Figure 1) revealed significant differences, with 7 proteins elevated in Cluster 2 compared to Cluster 1 (NOS3, PON2, MAP4K5, AIFM1, NUB1, STX8, and PDGFC) and 1 decreased protein (HPGDS). These proteins are involved in various metabolic pathways, with MAP4K5 being relevant to the interferon pathway, while PON2, AIFM1, and NUB1 are implicated in the regulation of apoptosis, a dysregulated process in SSc.
Conclusion: These findings suggest the presence of distinct clinical, serological, and proteomic clusters, indicating heterogeneous subgroups within SSc. The proteomic analysis also identifies potential biomarkers associated with organ damage, providing key insights into the underlying mechanisms of the disease. This stratification could be instrumental in tailoring patient management, predicting disease progression, and guiding targeted therapeutic strategies.
Clinical and Serological Characteristics of Patients by Cluster. ¶ stand for variables used in the clustering algorithm.
Score Plot of Proteins Identified by Cluster.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (