

Background: Monocytes have been identified as playing a significant role in the pathogenesis of systemic sclerosis (SSc), with CD16+ non-classical monocytes being particularly involved. Interferon type I and II pathways are known to play a significant role in the pathogenesis of systemic sclerosis (SSc). Therapies targeting type I interferon signaling are currently being explored as potential treatment options for patients with SSc.
Objectives: To characterize the transcriptomic differences of monocytes in active SSc patients compared to healthy controls.
Methods: Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation from 30 active SSc (ACR/EULAR 2013 criteria) patients and 10 age matched healthy controls (HC). SSc was defined as active based on the requirement to initiate immunosuppressive therapy or adjust of on-going immunosuppressive regimens due to disease progression (i.e. skin and lung fibrosis progression). Single-cell RNA sequencing was performed using a 10x Genomics droplet-based platform. Transcriptomic data were analyzed using the Seurat package and annotated for PBMCs subtypes. Cell proportions were analyzed with the Speckle R package. Differentially expressed genes (DEGs) were determined using the Seurat FindMarkers (MAST) with sex as a covariate to correct for sex-effects and applying the following thresholds: |log 2 FC| ≥ 0.5 and FDR ≤ 0.05. Subsequent pathway enrichment analysis was carried out using the clusterProfiler package. The AddModuleScore function was used to calculate the average expression of gene sets of interest.
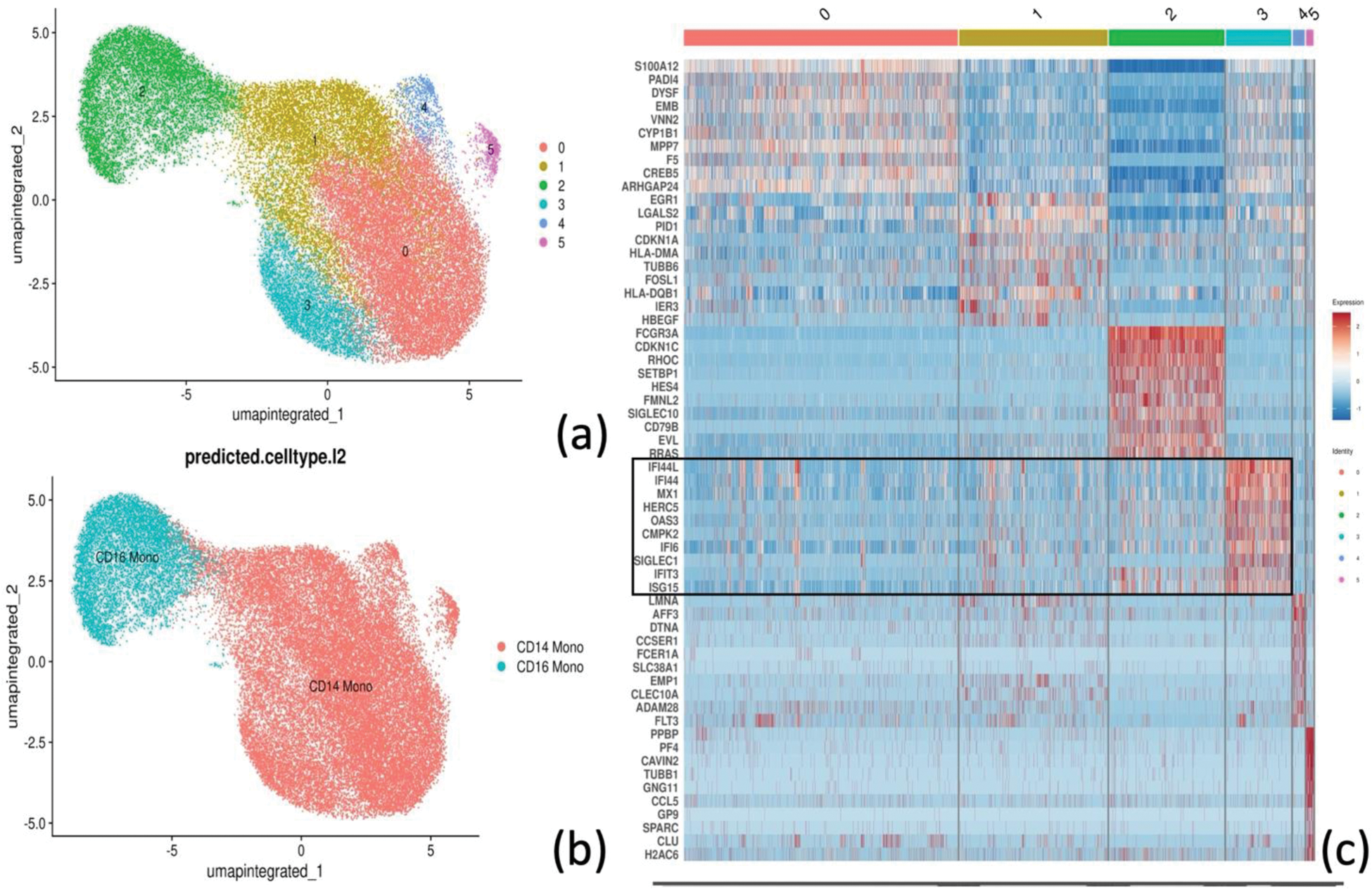
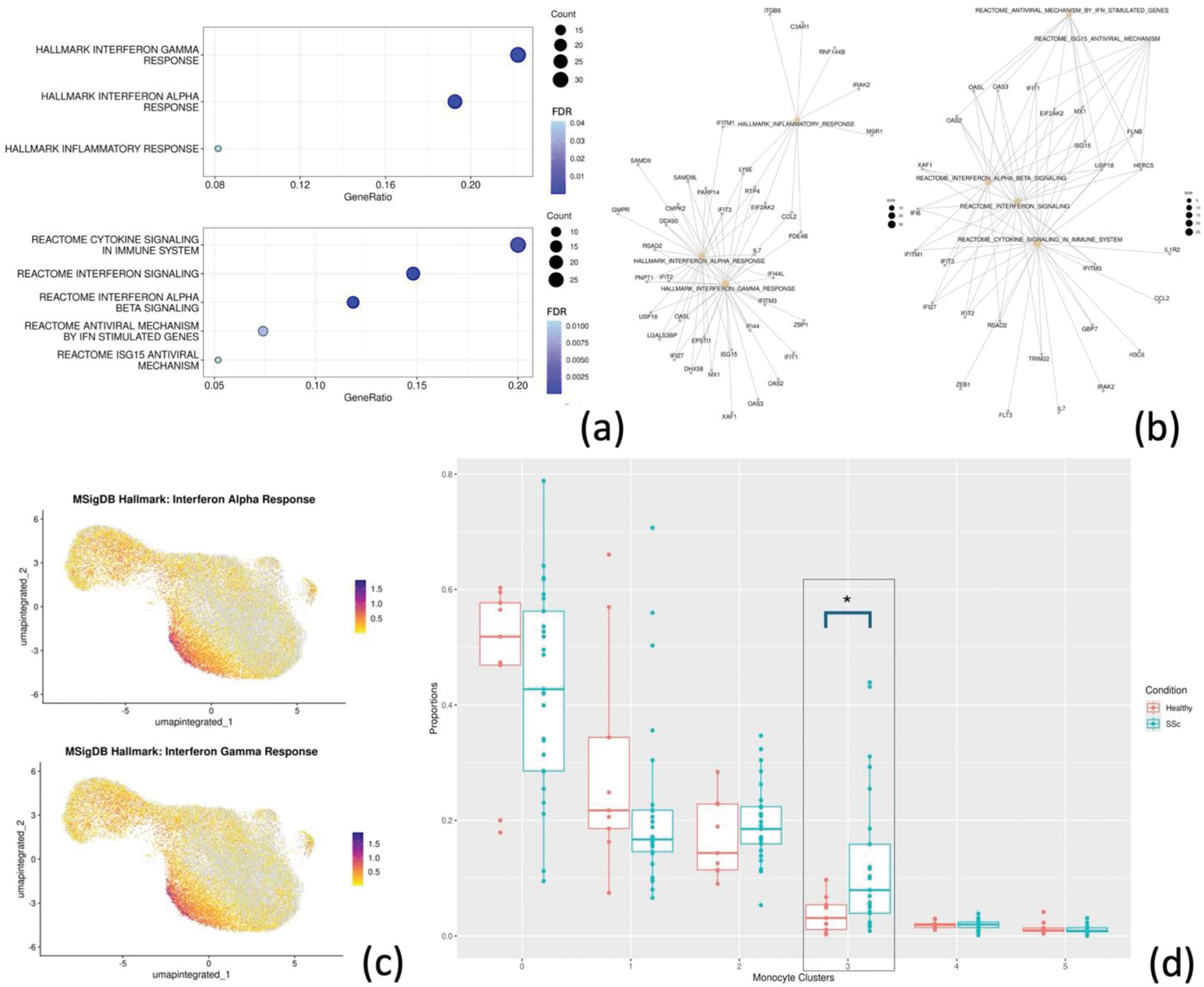
Results: Following PBMCs (N = 108,138) clustering and annotation from SSc patients and HC, monocytes (N = 38,071) were isolated, re-clustered (Figure 1a) and re-annotated in CD14+ and CD16+ (Figure 1b). Differential gene expression in monocytes yielded 186 upregulated and 233 downregulated DEGs in SSc compared to HC. Among upregulated DEGs, SSc monocytes were significantly enriched in interferon type I and II response genes (FDR < 0.01) (Figure 2a-b). Notably, among all monocyte clusters, CD14+ cluster 3 showed the highest expression of interferon response related genes (Figure 2c). This cluster expressed interferon-related genes (e.g. IFI44L, IFI44, SIGLEC1) among top marker genes (Figure 1c) and was significantly more abundant in SSc compared to HC (Proportion ratio 3.33, FDR 0.04, Figure 2d). Differential gene expression analysis between cluster 3 monocytes from SSc and HC identified 230 DEGs, including upregulation of genes related to monocyte extravasation and monocyte-to-macrophage transition, such as CX3CR1 (log 2 FC 0.54, FDR < 0.01) and ANPEP/CD13 (log 2 FC 0.59, FDR < 0.01).
Conclusion: In the peripheral blood of active SSc patients, monocytes exhibit an upregulated type I and type II interferon response gene signature, with CD14+ cluster 3 monocytes showing the strongest expression. This cluster, highly responsive to interferon as evidenced by its top marker genes, showed a significant three-fold increase in proportion in SSc. These findings confirm the role of monocytes in the pathogenesis of active SSc and suggest that CD14+ cluster 3 might be relevant for SSc tissue infiltration and inflammation.
Clustering (a), and annotation (b) of monocytes. Top 10 marker genes per each of the monocyte clusters (0 to 5), in the black box the marker genes for cluster 3 (c).
Pathway enrichment analysis represented with dot plot (a) and gene-concept network showing DEGs belonging to the different enriched pathways (b). Feature plot showing the interferon-alpha and -gamma response module score (c). Proportion analysis for the monocyte clusters (d).
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: Pietro Bearzi: None declared, Elena Pachera: None declared, Astrid Hofman: None declared, Lumeng Li: None declared, Laura Much: None declared, Kristina Buerki: None declared, Anna-Maria Hoffmann-Vold Boehringer Ingelheim, Janssen, Medscape, Merck Sharp & Dohme, Novartis and Roche, AbbVie, ARXX, Boehringer Ingelheim, Bristol Myers Squibb, Genentech, Janssen, Medscape, Merck Sharp & Dohme, Pliant Therapeutics, Roche and Werfen, Boehringer Ingelheim, Janssen, Mike O. Becker GSK, Amgen, Novartis, Vifor, Amgen, Vifor, Grant/research support from: Foundation for research in Rheumatology (FOREUM), EMDO Foundation, Novartis foundation for medical-biological research, Innovative Medicines Initiative (IMI), Roberto Giacomelli Astrazeneca, Alexion, Novartis, GSK, MSD, BMS, Boehringer-Ingelheim, Amgen, Abbvie, Roche, Galapagos, AnaptyBio, Argenx, Biogen, Ns-pharma, Johnson & Johnson, Astrazeneca, Alexion, Novartis, GSK, MSD, BMS, Boehringer-Ingelheim, Amgen, Abbvie, Roche, Galapagos, AnaptyBio, Argenx, Biogen, Ns-pharma, Johnson & Johnson, Abbvie, Johnson & Johnson, Oliver Distler 4P-Pharma, Abbvie, Acceleron, Acepodia Biotech, Aera, Alcimed, Altavant, Amgen, AnaMar, Anaveon AG, Argenx, AstraZeneca, Blade, Bayer, Boehringer Ingelheim, Calluna (Arxx), Cantargia AB, Catalyze Capital, Corbus, CSL Behring, Galderma, Galapagos, Glenmark, Gossamer, Horizon, Janssen, Kymera, Lupin, Medscape, MSD Merck, Miltenyi Biotec, Mitsubishi Tanabe, Nkarta Inc., Novartis, Orion, Pilan, Prometheus, Quell, Redxpharma, Roivant, EMD Serono, Topadur and UCB, 4P-Pharma, Abbvie, Acceleron, Acepodia Biotech, Aera, Alcimed, Altavant, Amgen, AnaMar, Anaveon AG, Argenx, AstraZeneca, Blade, Bayer, Boehringer Ingelheim, Calluna (Arxx), Cantargia AB, Catalyze Capital, Corbus, CSL Behring, Galderma, Galapagos, Glenmark, Gossamer, Horizon, Janssen, Kymera, Lupin, Medscape, MSD Merck, Miltenyi Biotec, Mitsubishi Tanabe, Nkarta Inc., Novartis, Orion, Pilan, Prometheus, Quell, Redxpharma, Roivant, EMD Serono, Topadur and UCB, Boehringer Ingelheim, Kymera, Mitsubishi Tanabe, UCB.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (