

Background: Despite advances in the treatment of rheumatoid arthritis (RA), optimal disease control cannot be achieved in approximately 5-20% of RA patients, and symptoms, particularly chronic pain, persist. This population presents a challenge in everyday clinical practice, thus referred to as difficult-to-treat (D2T). D2T RA is a highly heterogeneous group, with several contributing factors such as treatment-resistant illness, presence of comorbidities, secondary joint damage, and central pain sensitization; however, the exact pathomechanisms remain unclear. Peripheral blood mononuclear cells (PBMC) have been reported to reflect pathophysiological changes in the central nervous system (CNS) and, together with metabolomic analysis, have proven to be a valuable tool for investigating systemic inflammatory diseases.
Objectives: Our primary objective was to analyze the PBMC transcriptomic and plasma metabolomic profiles of RA patients in order to identify mechanisms of therapy resistance and connections between pain and inflammation.
Methods: A total of 28 D2T, 16 non-D2T clinically well-characterized RA patients, and 29 HCs were included in our study from two centers (Pécs and Budapest). Transcriptomic analysis was conducted using total RNA isolated from PBMCs through next-generation sequencing, and metabolomic profiling was obtained from blood plasma using the Biocrates MxP Quant 500 kit. The results were evaluated with bioinformatics tools.
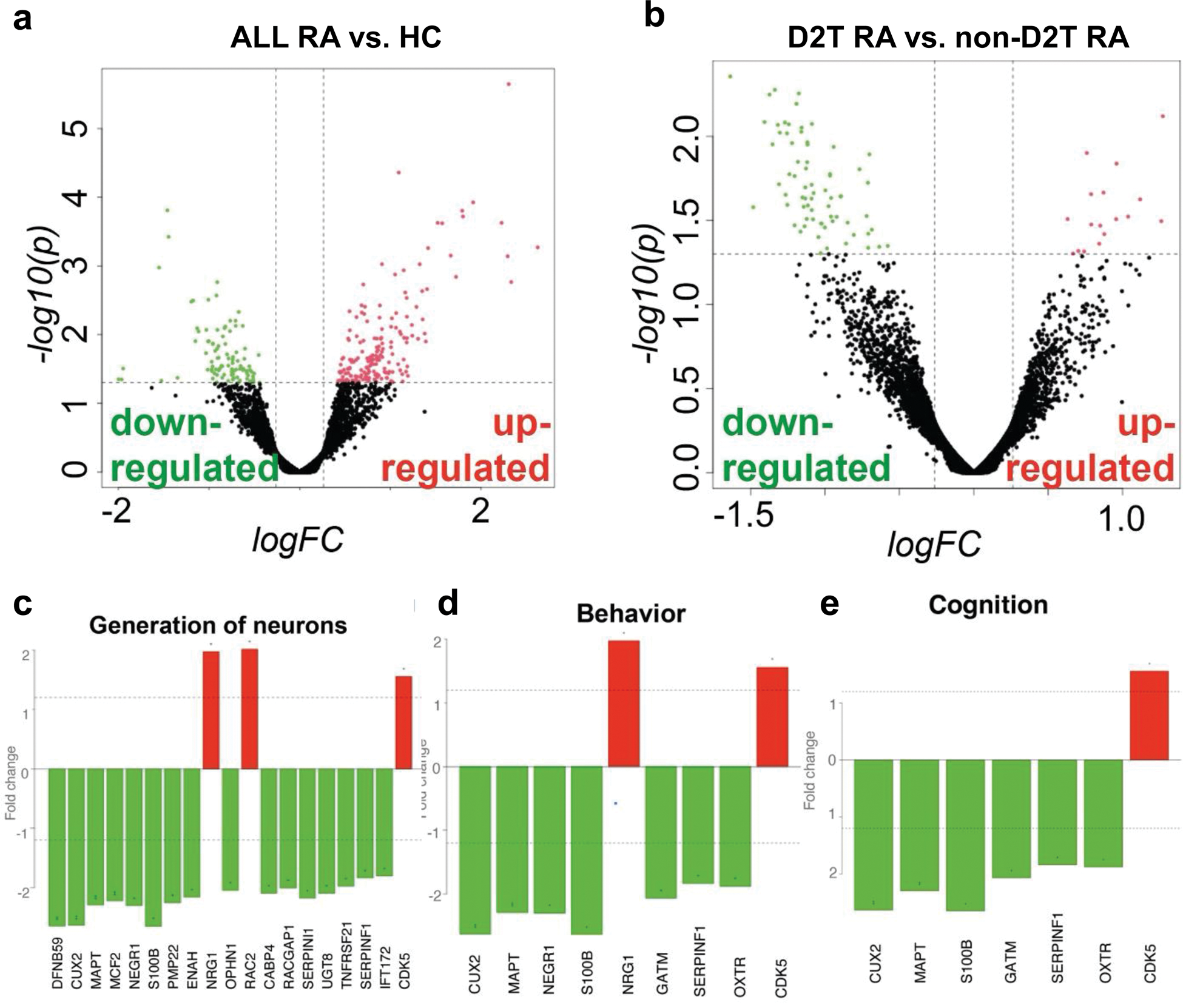
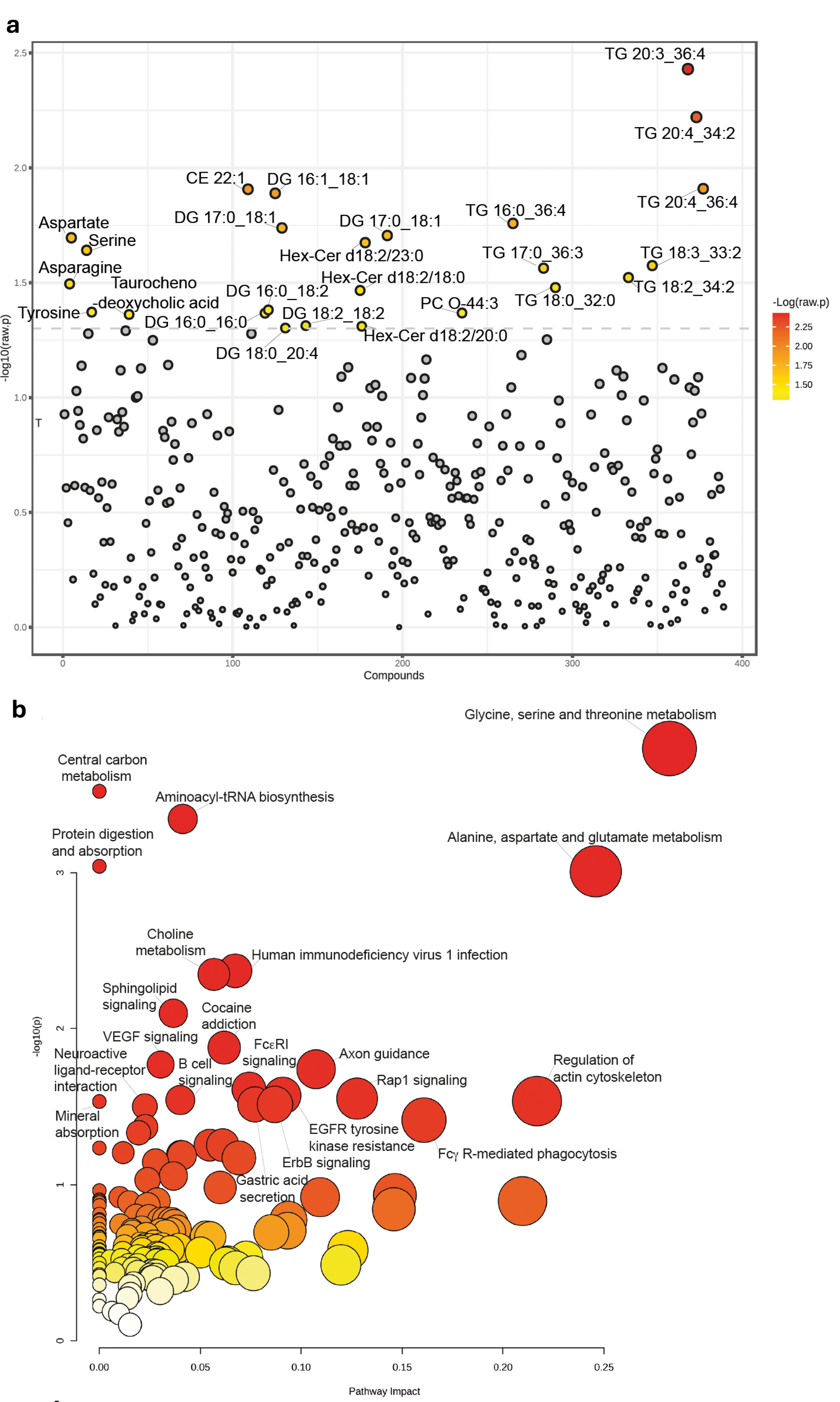
Results: All RA patients exhibited significantly different transcriptomic profiles compared to HCs. A comparison of all RA samples to HCs revealed 269 differentially expressed (DE) genes, consisting of 174 that are upregulated and 95 that are downregulated in RA (Figure 1a). Highly implicated genes include HBB (hemoglobin beta, p<0.001), LTK (leukocyte receptor tyrosine kinase, p=0.003), TLR4 (Toll-like receptor 4, p=0.005), and CXCL5 (chemokine ligand 5, p=0.035). Functional enrichment analysis using the Gene Ontology (GO) and Reactome databases explored the biological functions of DE genes in the RA group, revealing their roles in immune processes, oxygen transport, cell signaling, and communication. Comparing D2T to non-D2T RA patients, a total of 87 DE genes were identified, of which 17 were upregulated (e.g., NRG1, neuregulin-1 p=0.015) and 70 downregulated (e.g., NEGR1, neuronal growth regulator 1, p=0.017 or S100B encoding a calcium-binding protein mainly expressed in the CNS, p=0.026) in the D2T group (Figure 1b). Functional enrichment analysis of DE genes demonstrated the involvement CNS processes (generation of neurons (p<0.001), memory (p<0.001), cognition (p<0.001), and behavior (p<0.001) (Figure 1c-e). Metabolomic profiling revealed a group of lipids and metabolites that distinguish the D2T population from non-D2T, including several di- and triacylglycerols and amino acids, such as aspartate, serine, asparagine, and tyrosine (Figure 2a). Joint analysis of transcriptomic and metabolomic results further supported the complex interplay between inflammatory (e.g., sphingolipid metabolism, B-cell signaling) and neuronal functional processes (e.g., neuroactive-ligand receptor interaction, axon guidance) (Figure 2b).
Conclusion: Our analysis identified a characteristic transcriptomic and metabolomic profile in D2T RA patients. The transcriptomic analysis revealed many genes, including NRG1, NEGR1, and S100B, while the metabolomic analysis demonstrated many metabolites and lipids that may be associated with neuroinflammatory, neuroplastic, and neurodevelopmental processes. Together, these findings suggest the involvement of CNS processes and central sensitization in the development of the D2T state in RA. Our results may improve the understanding of therapy resistance and identify potential targets for novel treatments.
Volcano plot of transcriptomic analysis of all RA vs. HC (a) and D2T RA vs. non-D2T RA patients (b); Gene ontology (GO)-based enrichment analysis reveals significantly different pathways in the comparison of D2T RA vs. non-D2T RA (c-e).
Significantly different metabolites and lipids in the comparison of D2T vs non D2T RA patients (a) univariate statistics (t-test, p (raw)< 0.05); Joint pathway analysis based on significantly different metabolites and mRNAs (b) CE, cholesterol ester; DG, diglydercide; Hex-Cer, Hexosylceramides; PC, phosphatidylcholine; TG, triglyceride.
REFERENCES: NIL.
Acknowledgements: HUN-REN–PTE Chronic Pain Research Group (14017), OTKA K138046 and K131479, TKP2021-EGA-29, National Brain Research Program, Richter Gedeon PhD Scholarship (LGT).
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (