

Background: Biologics have transformed the management of axial spondyloarthritis (axSpA). However, their high cost and potential side effects preclude their use as first-line therapy. Currently, it is challenging to identify patients who would benefit from early biologics, resulting in trial-and-error use of NSAIDs followed by a response evaluation over months before initiating biologics. In South Korea, insurance guidelines mandate at least three months of treatment with two NSAIDs or csDMARDs before biologics can be prescribed, causing a delay of 3–6 months for patients who may not respond to first-line therapies. This delay subjects these patients to unnecessary pain and disease progression, highlighting the need for predictive tools to identify those who would benefit from early biologics treatment.
Objectives: This study aimed to characterize patients requiring early biologics through single-cell RNA sequencing (scRNA-seq) analysis of peripheral blood mononuclear cells (PBMCs) obtained from newly diagnosed axSpA patients.
Methods: PBMCs were collected at baseline from axSpA patients newly diagnosed at Samsung Medical Center. Patients initiating biologics within six months were categorized as early biologics users, while those who did not were categorized as non-early biologics users. Single-cell RNA sequencing data were processed using Cell Ranger, and quality control was performed using Seurat. Cell types were annotated using external reference datasets from ankylosing spondylitis (AS) studies [1]. Differences in cell proportions were analyzed with scProportionTest, and differentially expressed genes (DEGs) were identified using the Wilcoxon Rank Sum test. Pathway enrichment analyses were conducted using KEGG and GO terms.
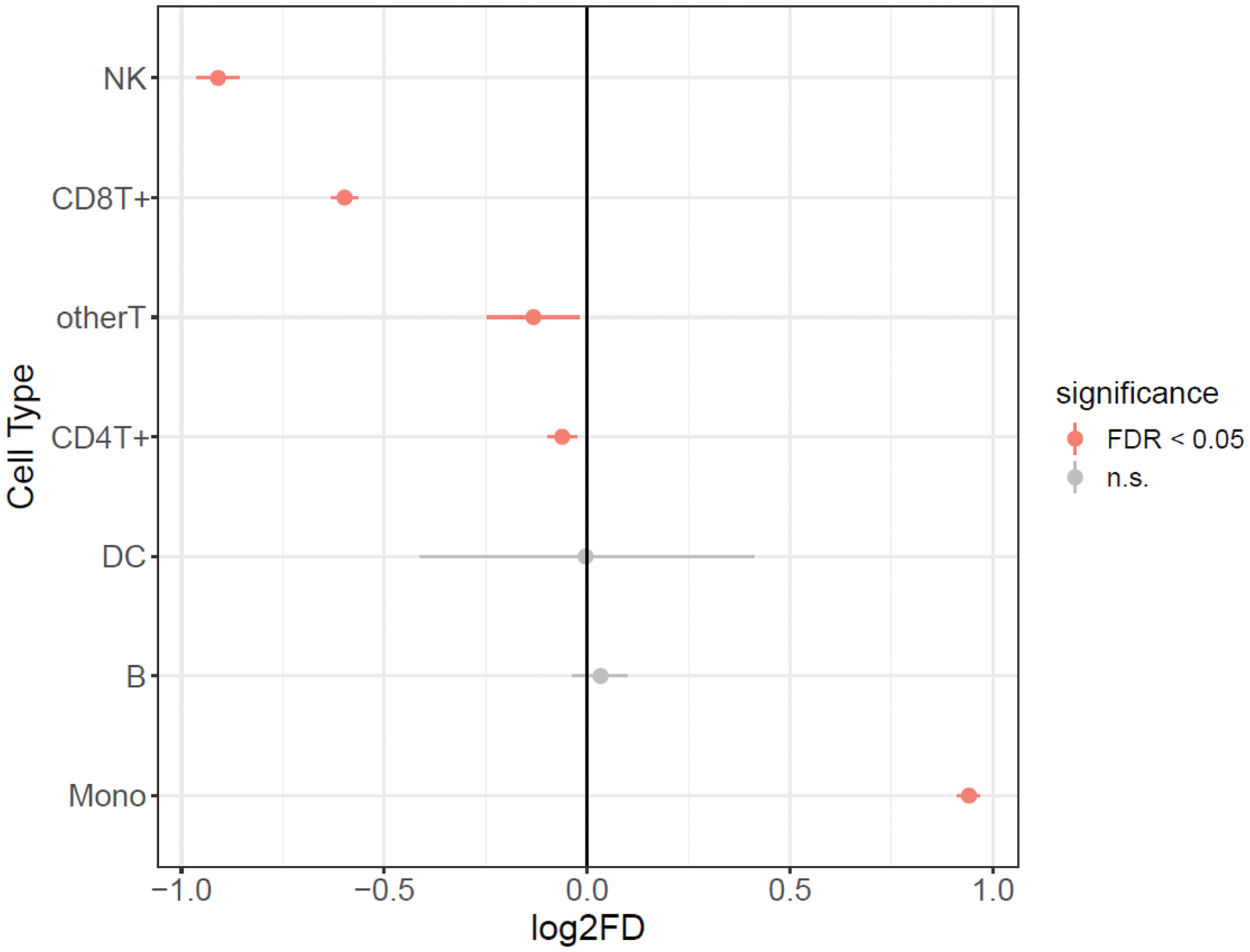
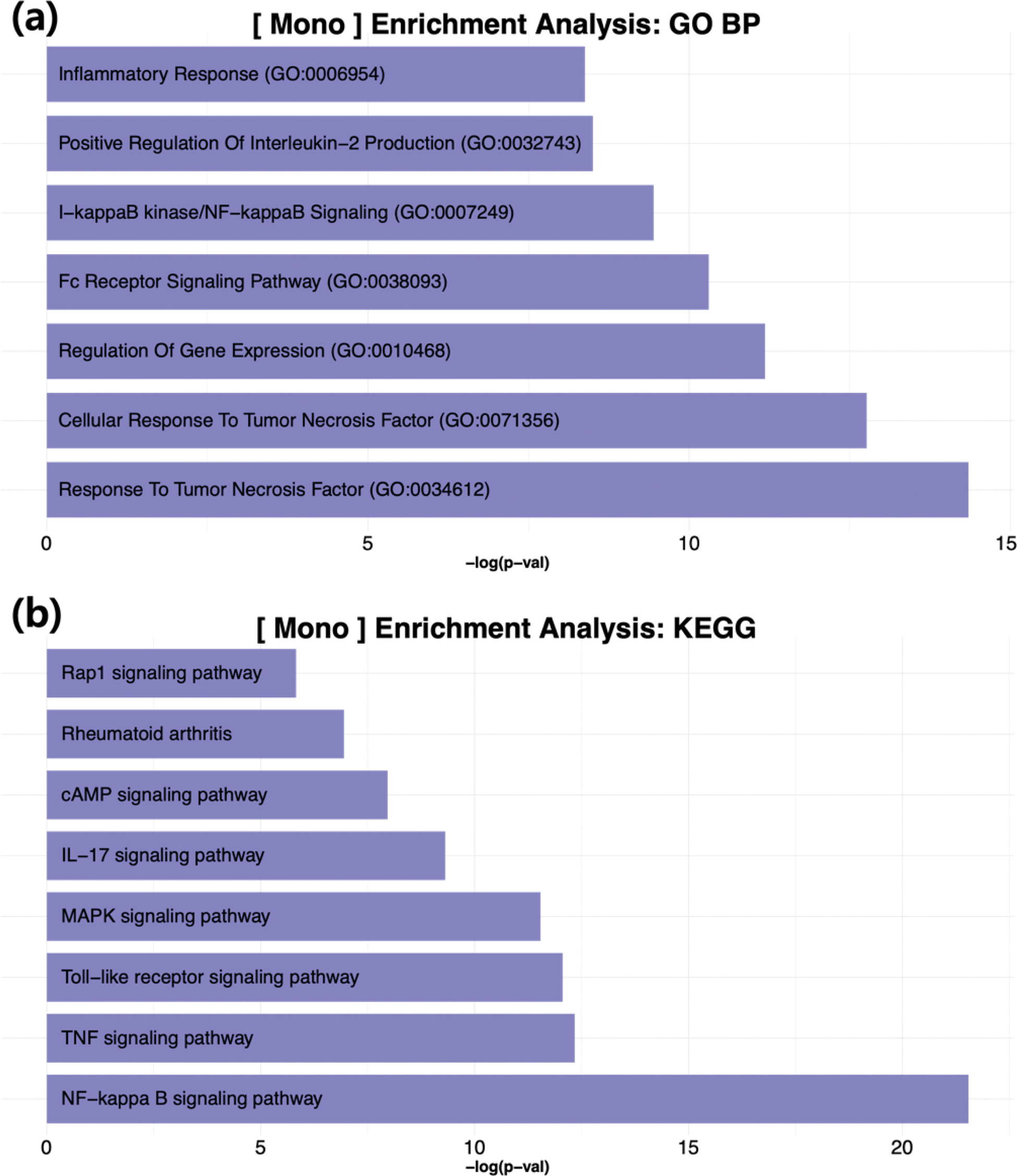
Results: Nineteen patients with axial spondyloarthritis (axSpA) were enrolled, of whom seven were categorized as early biologics users (initiating biologics within six months) and twelve as non-early biologics users. A total of 157,029 single cells passed quality control and were included in subsequent analyses. Differential cell proportion analysis revealed marked differences between early and non-early biologics users. Specifically, early biologics users exhibited a significant increase in monocyte proportions, along with a decrease in NK and CD8+ T cells, suggesting an altered balance in both innate and adaptive immunity. Monocytes had the highest number of differentially expressed genes (DEGs), followed by CD8+ T cells and NK cells, highlighting the potential importance of monocytes in determining early treatment responses (Figure 1). Enrichment analyses of monocyte-specific DEGs indicated significant activation of pathways closely linked to the pathogenesis of axSpA, including NF-κB signaling, the Toll-like receptor signaling pathway, the response to tumor necrosis factor, and MAPK signaling [2] (Figure 2). Within the I-kappaB kinase/NF-kappaB signaling pathway, genes such as NFKBIA, TBK1, IRAK2, RIPK2, REL, TANK, NFKB1, CARD11, RELB, and BIRC3 overlapped in the GO Biological Process gene set, aligning with previous research demonstrating that canonical and noncanonical NF-κB pathways promote monocyte differentiation into bone-resorbing osteoclasts [3]. In CD8+ T cells, DEGs were enriched in the HIF signaling pathway, with genes including LDHB, IFNGR2, BCL2, CYBB, PRKCA, IL6R, IGF1R, and PDK1 overlapping in the KEGG gene set. These findings are consistent with previous reports emphasizing the necessity of increased glycolysis in supporting CD8+ T cell activation and differentiation [4]. Similarly, NK cells in early biologics users displayed differential expression of genes such as FPR1, XCL2, CYBB, CD14, FOS, LYZ, S100A9, AIF1, and S100A8, which are enriched in inflammatory response pathways. NK cells have been implicated in several immune-mediated diseases [5], and these results suggest that their dysregulation may contribute to the early disease process in axSpA.
Conclusion: These data indicate that early biologics users exhibit a distinct immunological profile characterized by enhanced monocyte-driven inflammation and concurrent alterations in CD8+ T cell and NK cell populations. These findings underscore the critical role of innate immunity, particularly through monocyte-mediated pathways, in identifying patients who may benefit from early biologics therapy, offering a basis for personalized therapeutic strategies.
REFERENCES: [1] Liu J, et al. Combined Single Cell Transcriptome and Surface Epitope Profiling Identifies Potential Biomarkers of Psoriatic Arthritis and Facilitates Diagnosis via Machine Learning. Front Immunol . 2022;13:835760.
[2] Assassi S, et al. Whole-blood gene expression profiling in ankylosing spondylitis shows upregulation of toll-like receptor 4 and 5. J Rheumatol . 2011;38(1):87-98.
[3] Baum R, et al. Bone as a target organ in rheumatic disease: impact on osteoclasts and osteoblasts. Clin Rev Allergy Immunol 2016; 51: 1–15.
[4] Doedens, Andrew L., et al. “Hypoxia-inducible factors enhance the effector responses of CD8+ T cells to persistent antigen.” Nat immunol 14.11 (2013): 1173-1182.
[5] Conigliaro P, et al. Emerging role for NK cells in the pathogenesis of inflammatory arthropathies. Autoimmun Rev . 2011;10(10):577-581.
Single-cell proportion test results.
To compare cell type proportions between early and non-early users, a single-cell proportion test was performed using the R package scProportionTest.
Results of the enrichment analysis.
Enrichment analyses using (a) GO Biological Process and (b) KEGG gene sets.
Acknowledgements: This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: RS-2024-00439690).
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (