

Background: Rheumatoid arthritis (RA) is driven by the activation of complex inflammatory pathways involving immune cell infiltration into synovial tissue and fluid. Synovial fluid (SF) offers valuable insights into the pathophysiological mechanisms of joint disease. Few studies have applied advanced multiplex or single-cell transcriptomic techniques to analyze its cytokine and cellular composition.
Objectives: This study aimed to: (1) characterize the cytokine profile of SF in RA compared to other chronic inflammatory rheumatic diseases (CIRD) and osteoarthritis (OA); (2) compare cytokine levels in SF to paired serum samples in RA patients; and (3) explore the single-cell transcriptomic signature of RA SF mononuclear cells (SFMCs) compared to peripheral blood mononuclear cells (PBMCs).
Methods: SF samples were collected from 115 patients for cytokine analysis: 45 with RA, 48 with other CIRD (9 psoriatic arthritis, 12 spondyloarthritis, 14 unclassified CIRD, 13 juvenile idiopathic arthritis), and 22 with OA. Among these 115 patients, paired serum samples were available for 29 RA patients. Levels of 21 cytokines were measured in SF and serum using Luminex technology, including TNF-α, IL-1β, IL-6, IL-17, GM-CSF, IL-23, and CCL20. Single-cell RNA sequencing (scRNA-seq) was performed on paired SFMCs and PBMCs from six RA patients using 10X Chromium technology. Data were extracted using CellRanger pipelines. Differences in cellular composition were compared using fold change (FC) analysis with Monte Carlo simulations.
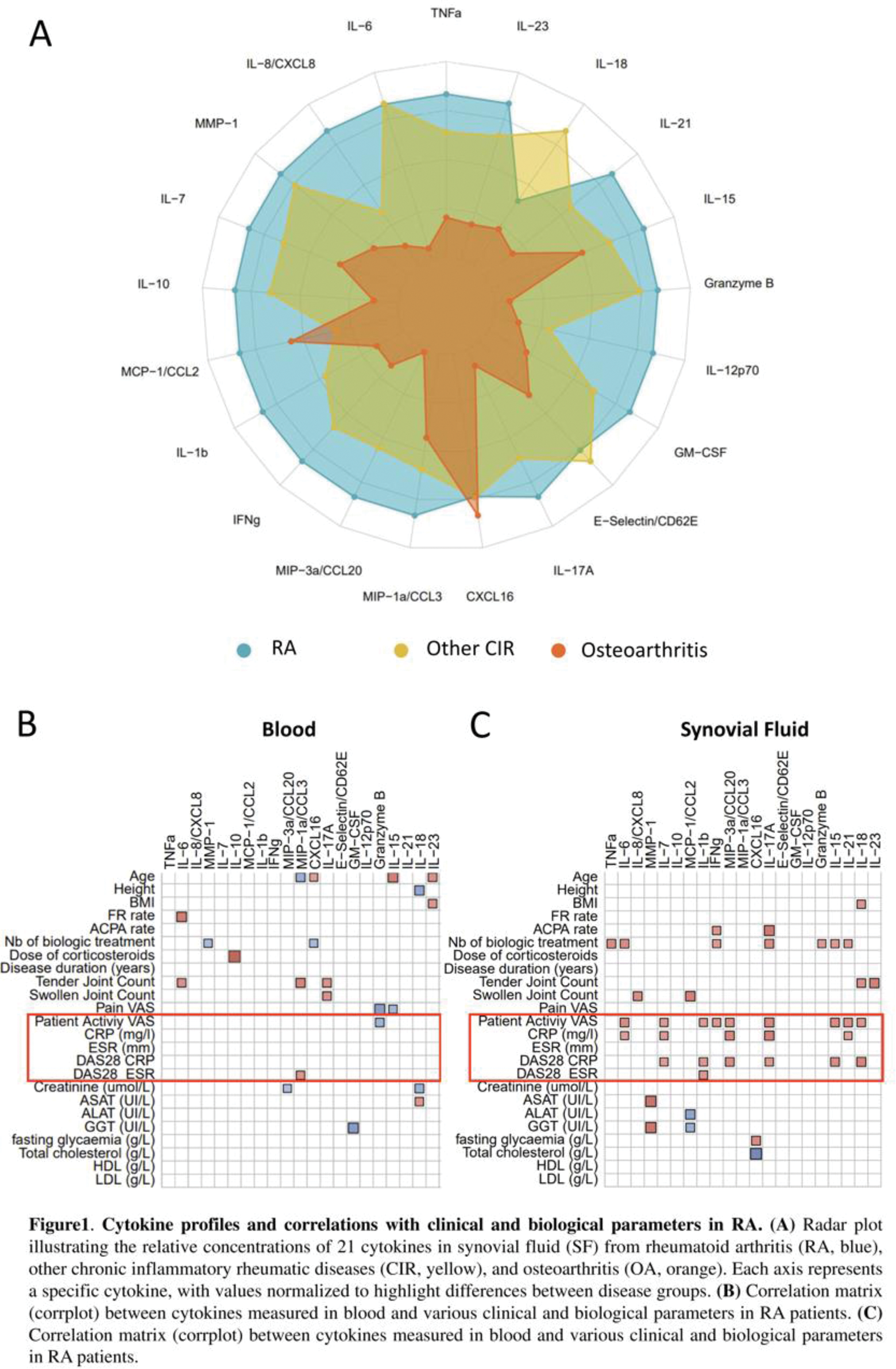
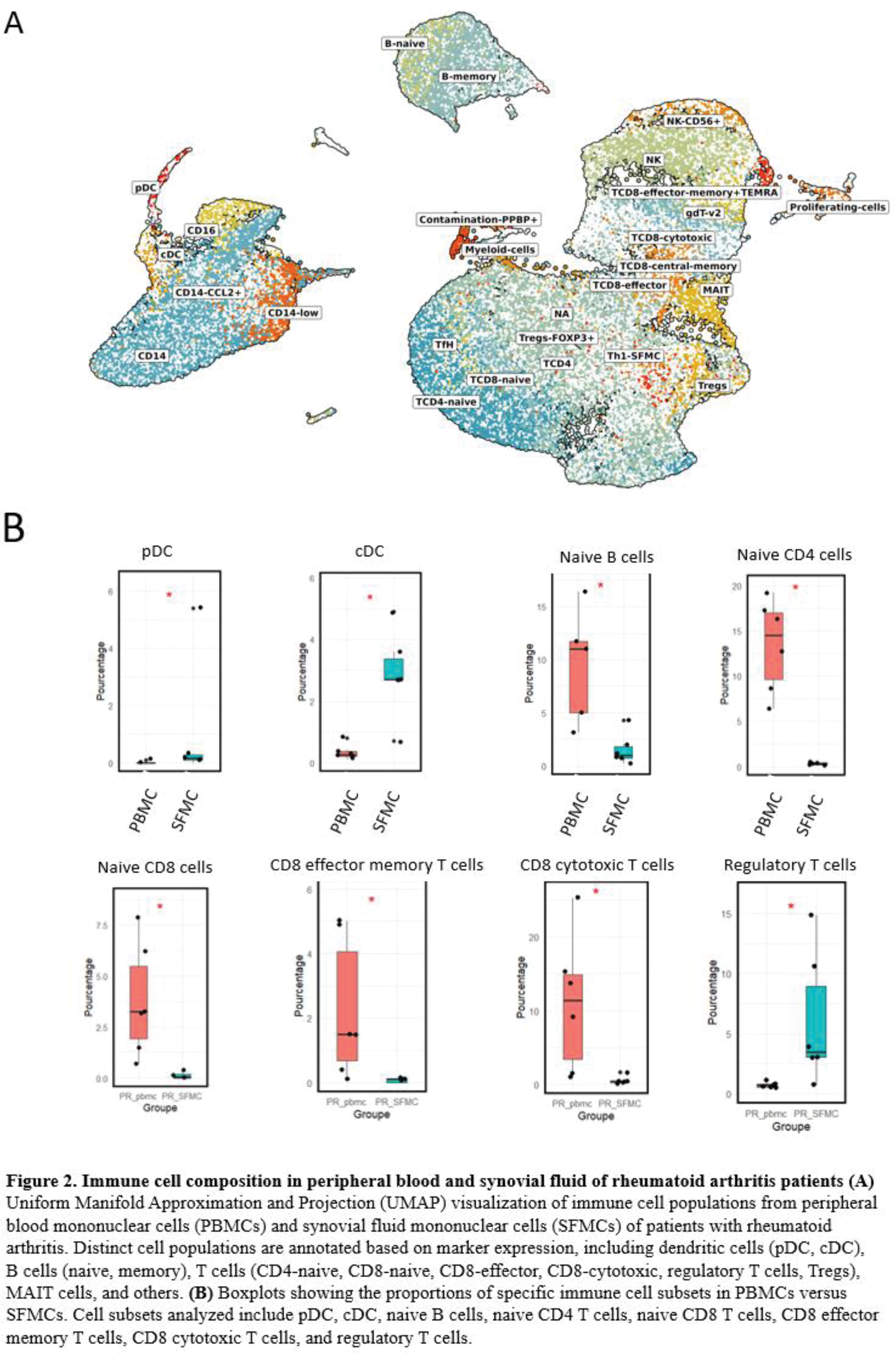
Results: Cytokine analysis revealed significantly elevated levels of pro-inflammatory mediators in RA SF compared to OA and CIRD. Notably, TNF-α, IL-1β, IL-6, IL-17, Granzyme B, and CCL20 were highly expressed in RA SF compared to OA (Figure 1A). Additionally, IL-8, CCL2, and CCL3 were significantly increased in RA SF compared to CIRD. Interestingly, heatmap clustering of RA cytokine profiles revealed two distinct patient subgroups. One group exhibited high cytokine levels, associated with more inflammatory SF (increase of leucocyte number and %PNN) and elevated CRP levels, while the other displayed a less pronounced inflammatory profile. The comparison of cytokine levels in paired RA SF and serum samples revealed significantly higher concentrations of inflammatory cytokines in SF. Moreover, cytokines in SF were strongly correlated with clinical activity markers (CRP, DAS28-CRP), whereas no significant correlations were observed in serum (Figure 1B-C). To investigate the cellular drivers of synovial inflammation, single-cell RNA sequencing (scRNA-seq) was performed on 6 paired SFMCs and PBMCs. This analysis revealed 27 distinct cellular subpopulations, encompassing 7 myeloid clusters and 20 lymphoid clusters (Figure 2A). Among these, SFMCs were enriched in conventional dendritic cells (DC) (FC = 3.5, p = 0.028) as well as plasmacytoid DC (FC = 5.5, p = 0.028) (Figure 2B). In contrast, naive lymphocytes were depleted in SF, together with decreased B cells (FC=-2.17, p=0.028), CD4 T cells (FC = -5.9, p=0.028) and CD8 T cells (FC = -6.5, p=0.028) (Figure 2B). A distinct population of CD4+ T cells expressing PD-1, Granzyme A, IL-17, and IFN-γ was identified exclusively in SFMCs, indicative of a highly pro-inflammatory cell subset. This was accompanied by reduced levels of cytotoxic CD8+ T cells (FC = -4.9, p = 0.028) and effector memory CD8+ T cells (FC = -5.2, p = 0.028) in SFMCs (Figure 2B). Additionally, regulatory T cells were enriched in SF compared to blood (FC = 2.9, p = 0.028) (Figure 2B).
Conclusion: This study emphasizes the distinctive inflammatory milieu of RA synovial fluid (SF), distinguished by elevated cytokine levels and immune cell shifts when compared to serum and other rheumatic diseases. Luminex analysis identified two distinct cytokine profiles in SF, underscoring its heterogeneity and significant correlation with clinical markers, which are absent in serum. This finding reinforces SF as a pivotal site of inflammation. Single-cell transcriptomics revealed a mature immune response in SF mononuclear cells (SFMCs), with dendritic cell enrichment, naive lymphocyte depletion, and elevated levels of CCL20, CCL3, CCL2, and CCL8 in Luminex analysis, driving dendritic cell activation and naïve T cell recruitment. Increased levels of Granzyme B and IL-17 align with CD4 T cell transcriptomic signatures (PRF1, IL-17A, GZMA), indicating their role in joint inflammation. This immune imbalance, characterized by a cytotoxic CD4 T lymphocyte response, reduced CD8 T cells, and enriched regulatory T cells, highlights novel pathways in RA pathophysiology and synovial compartment targeting.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (