

Background: Systemic lupus erythematosus is an autoimmune disease characterized by autoantibody production and deposition of immune complexes. SLE patients are exposed to an increased cardiovascular risk, mainly by atherosclerosis, increasing their morbidity and mortality [1]. Although the presence of traditional cardiovascular risk factors contributes to this process, they cannot fully explain the increased cardiovascular risk in patients with SLE [3]. Immune mechanisms underlying chronic inflammation, immune dysfunction, and endothelial injury are regarded to accelerate the atherosclerotic process in SLE [2]. Atherosclerosis in SLE is characterized by increased carotid intima-media thickness as determined by carotid ultrasound, a surrogate marker of subclinical atherosclerosis [4]. This study integrates transcriptomic data from SLE subjects with elevated CIMT and immune cell profiling in peripheral blood to identify key molecular and cellular pathways underlying SLE-related atherosclerosis.
Objectives: This study aimed to identify molecular mechanisms and immune cell patterns associated with atherosclerosis in SLE patients.
Methods: Transcriptomic datasets GSE154851 and GSE110174 were sourced from the Gene Expression Omnibus platform. GSE154851 includes data from 38 patients suffering from SLE with carotid ultrasound-verified increased CIMT, and GSE110174 includes 144 SLE patients with no increased CIMT. Both included one healthy control group, denoted as HC. Gene Set Enrichment Analysis was performed with the aim of exploring enriched genes associated with coronary atherosclerosis. PPI networks were constructed by Cytoscape and HGs were screened by five different algorithms: DMNC, Degree, Stress, Radiality, Eccentricity. Immune cell composition and abundance for 22 immune cell types were analyzed by CibersortX. A two-way ANOVA with the Benjamini, Krieger, and Yekutieli multiple comparisons test was used to find the difference between the three groups. Logistic regression identified immune cell type associated with SLEat state. A p < 0.05 was considered statistically significant.
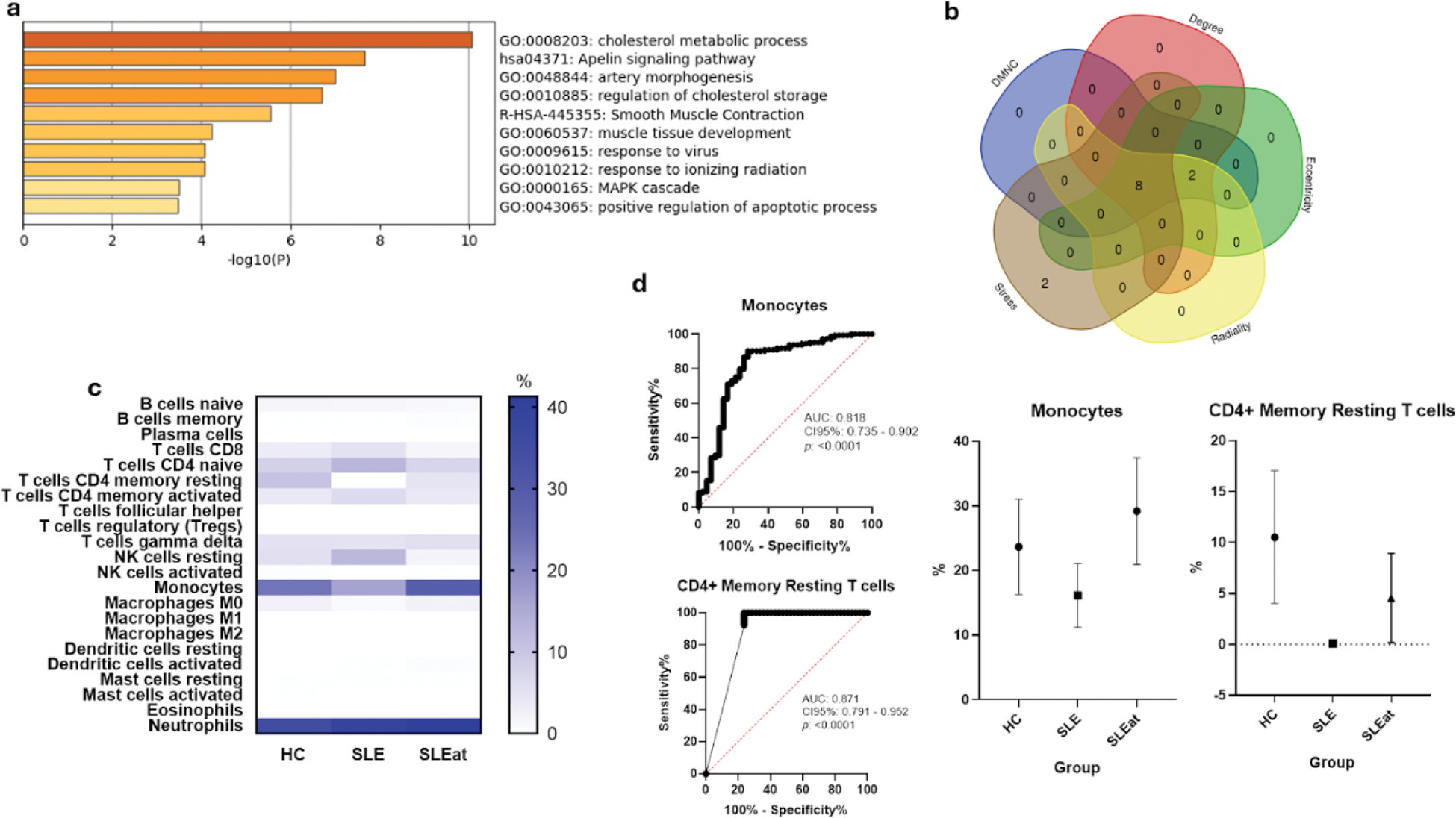
Results: 19 genes were enriched in the coronary atherosclerosis phenotype. Figure 1a presents the top functional categories, and the overlapping hub genes identified using five algorithms were ACTA2 , ABCA1 , MYH11 , APOB , PCSK9 , PPARG , TGFBR1 , and SMAD3 (Figure 1b). Proportions of CD8+ T cells, CD4+ naïve T cells, CD4+ memory resting T cells, CD4+ memory activated T cells, resting NK cells, monocytes, M0 macrophages, and neutrophils, as shown in Figure 1c, were significantly different among groups. Logistic regression analysis revealed that CD8+ T cells, CD4+ naïve T cells, CD4+ memory activated T cells, and resting NK cells were negatively related to SLEat, while CD4+ memory resting T cells, monocytes, and M0 macrophages did positively. The details are shown in Table 1, and further confirmed in a ROC curve analysis for diagnostic potential, as indicated in Figure 1d.
Immune cell composition and diagnostic potential of the hub genes in SLE-associated atherosclerosis. (a) Enrichment analysis of genes associated with coronary atherosclerosis in SLE patients. (b) Overlapping hub genes identified by the five different algorithms. (c) Proportions of 22 different immune cell types across healthy controls (HC), SLE patients without CIMT (SLE), and SLE patients with increased CIMT (SLEat). (d) ROC curves representing the diagnostic value of monocytes and CD4+ memory resting T cells for atherosclerosis in SLE patients.
Logistic Regression Analysis of Immune Cell Types Associated with SLE-Related Atherosclerosis.
| Cell Type | OR | CI95% | p |
|---|---|---|---|
| CD8+ T cells | 0.72 | 0.62 - 0.85 | <0.001 |
| CD4+ Naïve T cells | 0.81 | 0.74 - 0.89 | <0.001 |
| CD4+ Memory Resting T cells | 4.81 | 2.48 - 9.35 | <0.001 |
| CD4+ Memory Activated T cells | 0.36 | 0.25 - 0.51 | <0.001 |
| Resting Natural Killer cells | 0.27 | 0.14 - 0.50 | <0.001 |
| Monocytes | 1.41 | 1.26 - 1.58 | <0.001 |
| Macrophages M0 | 2.71 | 1.87 - 3.91 | <0.001 |
Odds ratios (OR), confidence intervals (CI95%), and p-values for immune cell types positively or negatively associated with SLE patients with increased carotid intima-media thickness (SLEat) compared to SLE patients without increased CIMT.
Conclusion: This work integrated immune cell abundance with transcriptomic data to unravel the mechanisms underlying atherosclerosis in SLE patients. We identified immune cell profiles associated with atherosclerosis, including positive associations with the following cell subsets: monocytes, CD4+ memory resting T cells, and M0 macrophages, while cells as CD8+ T cells and resting NK cells were negatively associated. These immune cell signatures are indicative of a special way through which SLE-related atherosclerosis occurs via an immunopathogenic pathway. These results underscore the value of personalized approaches in the management of cardiovascular risk in SLE patients and give further reason for research into validation in larger cohorts.
REFERENCES: [1] Bartoloni, E., Alunno, A., & Gerli, R. (2022). Cardiovascular disease risk in systemic lupus erythematosus and the role of traditional and non-traditional factors. Frontiers in Medicine , 9, 847635.
[2] Gkrouzman, E., Costenbader, K. H., & Gladman, D. D. (2021). Cardiovascular disease in systemic lupus erythematosus: Risk, prevention, and management. Rheumatic Disease Clinics of North America , 47(3), 401-416.
[3] Nikpour, M., Urowitz, M. B., & Gladman, D. D. (2010). Epidemiology of atherosclerosis in systemic lupus erythematosus. Current Rheumatology Reports , 12(4), 250-258.
[4] Sacre, K., Escoubet, B., Granger, B., et al. (2017). Accelerated atherosclerosis in systemic lupus erythematosus: A controlled study of preclinical atherosclerosis in 70 patients. Arthritis & Rheumatology , 69(7), 1277-1287.
Acknowledgements: We’d like to thank the Mexican National Council of Humanities, Science and Technology (CONAHCYT) for providing the academic grant that made this research possible. Grant ID: PCC-2022-320697.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (