

Background: Serpins are a superfamily of proteins, most of whose members were initially identified as protease inhibitors. Serpins have emerged as potential biomarkers and therapeutic targets in autoimmune diseases such as systemic lupus erythematosus (SLE) and lupus nephritis (LN). In particular, urinary SERPINA3 is a potential biomarker of response to treatment in patients with LN [1]. LN affects up to 50-60% of the patients with SLE and is one of the leading causes of morbidity and mortality in this population. Although some Serpins have been identified as regulators of inflammatory cascades, their exact role in the pathogenesis of SLE and LN is poorly understood. These serpins participate in various biological processes such as the regulation of endopeptidase activity, modulation of the extracellular matrix, and the regulation of complement and coagulation cascades.
Objectives: Here, we explored the expression and function of Serpins in patients with SLE and LN, and the utility of these biomarkers for disease activity evaluation and treatment response analyzing transcriptome data.
Methods: We obtained the microarray expression dataset GSE99967 from the Gene Expression Omnibus platform. This dataset contained the data of 29 patients with active LN, 13 patients with non-renal SLE, and 17 healthy controls. Median fluorescence intensity (MFI) of SERPIN expressions was analyzed. Linear regression with SLEDAI as a dependent variable was calculated, and logistic regression for the identification of associations with LN was carried out. The CibersortX algorithm calculated the composition and abundance of 22 types of immune cells (cell proportions) and their correlation was tested with the Serpins’ MFIs by using Spearman’s correlation test.
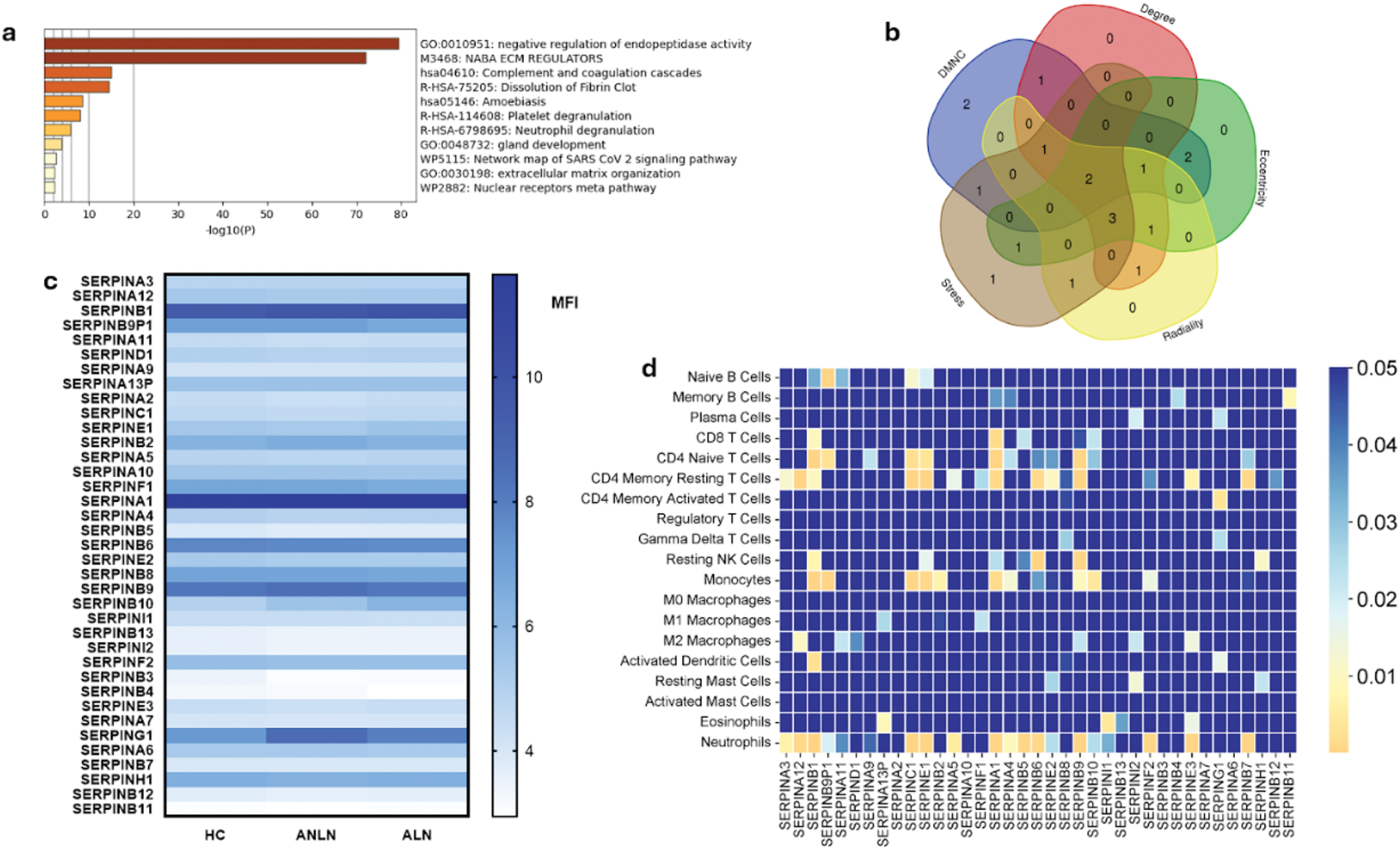
Results: Among the microarray data provided by Affymetrix Human Gene 2.0 ST Array, 37 genes from the Serpin family have shown involvement in negative regulation related to endopeptidase activity, extracellular matrix, complement, and coagulation cascades (Figure 1a). Among them, SERPINA3 and SERPING1 were consistently identified across five different algorithms. As compared between the three groups, significant differences were found in the MFIs of SERPINB10 , SERPING1 , and SERPINB3 , as depicted in Figure 1c. SERPINA3 showed significant association with disease activity when adjusting for LN class mixed 3 + 5, 4 + 5 and classes 4 and 5 (B coefficient: 0.743, p = 0.035, CI95%: 2.643 - 50.805), as well as for SLE patients who were taking antimalarials (B coefficient: 0.626, p : 0.001, CI95%: 7.314 - 25.238). On the counterpart, SERPINB9P1 was associated negatively with LN state (OR: 0.029, p : 0.012, CI95%: 0.002 - 0.463). Additionally, significant correlations were observed between several serpin proteins and immune cells, as shown in Figure 1d.
Functional Enrichment, Expression Profile, and Immune Cell Correlations of SERPIN Proteins in Lupus Nephritis . (A) Functional enrichment analysis shown biological process associated with SERPIN proteins, in particular negative regulation of endopeptidase activity, regulation of extracellular matrix and complement and coagulation cascades; (B) venn diagram showing the overlap of SERPIN proteins across five different algorithms used in the analysis; (C) heatmap of MFI values of SERPIN protein expression in HC, ALN and ANLN patients; (D) correlation matrix presenting correlations between SERPIN proteins and the proportions of different immune cell populations. Color describes the strength of statistical significance; blue means p > 0.05.
Conclusion: These data highlight that some Serpins, such as SERPINA3 and SERPINB9P1 correlates with disease activity and LN state, suggesting that those molecules may be employed as monitoring biomarkers and possible therapeutic targets in the treatment of SLE and LN. These results need to be confirmed prospectively, and their clinical use explored in further studies.
REFERENCES: [1] Araujo, E. N., Duarte, A. J. S., & da Silva, N. P. (2021). Urinary biomarkers in lupus nephritis: An update. Journal of Immunology Research , 2021, 6623451.
[2] Davidson, A., & Aranow, C. (2022). Pathogenesis and treatment of lupus nephritis. Current Opinion in Rheumatology , 34(3), 159-166.
[3] Silverman, G. A., Bird, P. I., Carrell, R. W., et al. (2018). The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Journal of Biological Chemistry , 293(11), 4035-4044.
[4] Tsokos, G. C. (2019). Systemic lupus erythematosus. New England Journal of Medicine , 381, 2111-2124.
Acknowledgements: We’d like to thank the Mexican National Council of Humanities, Science and Technology (CONAHCYT) for providing the academic grant that made this research possible. Grant ID: PCC-2022-320697.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (